For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSSTSALLDLDQFEKLAAAGEITTVICAAPDPYGRLVGKRLTISAFRTLGLDGGGVNASSFIFAVDLEMN PLDLPVSNAANGWVDIRLVPDLSTLRRVPWEPTAALVICDAYHMDSDEPLAVAPRTILRQQIDRAAQQGL RFKFASELEFYLSCTPPRQAWDLGYQNLKMMSDYRSDYQMIQAARDDWFIERIRNQMPEFGVPIESSKPE WGLGQQEVTLDYCDTLEMADRHVLFKYGVKQLAHSADLTVTFMAKPKIDEVGSSCHLHVSMWSTDDDQPL CWCPERRGMSDRFGAFVAGQLRHAIDMSLMYAPTVNSYKRFKPDQFAGTAIALGMDNRSCAFRLVGNGPS FRVENRIPGADVNPYYAYAATIAAGLDGVEANLPAPDIFDGNAWTREGVATVPTSLHRSLDLFAESKMAR TAFGDEVFDHLLSSAQGELEAFDSHTVTDWESRRYYERV
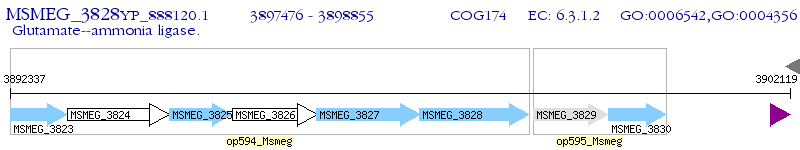
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3828 | - | - | 100% (459) | glutamine synthetase |
| M. smegmatis MC2 155 | MSMEG_2595 | - | 1e-92 | 42.21% (462) | gamma-glutamylisopropylamide synthetase |
| M. smegmatis MC2 155 | MSMEG_1116 | - | 7e-62 | 33.94% (442) | gamma-glutamylisopropylamide synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2885c | glnA4 | 6e-93 | 43.21% (449) | glutamine synthetase |
| M. gilvum PYR-GCK | Mflv_4065 | - | 3e-94 | 43.10% (464) | glutamate--ammonia ligase |
| M. tuberculosis H37Rv | Rv2860c | glnA4 | 6e-93 | 43.21% (449) | glutamine synthetase |
| M. leprae Br4923 | MLBr_01631 | glnA2 | 5e-38 | 33.08% (390) | glutamine synthase class II |
| M. abscessus ATCC 19977 | MAB_1920 | - | 1e-37 | 32.74% (394) | glutamine synthetase |
| M. marinum M | MMAR_3294 | glnA2 | 3e-37 | 31.44% (388) | glutamine synthetase GlnA2 |
| M. avium 104 | MAV_3718 | glnA | 6e-95 | 43.29% (462) | glutamine synthetase catalytic domain |
| M. thermoresistible (build 8) | TH_3396 | - | 6e-41 | 32.99% (394) | PUTATIVE glutamine synthetase, type I |
| M. ulcerans Agy99 | MUL_1335 | glnA2 | 1e-37 | 31.44% (388) | glutamine synthetase GlnA2 |
| M. vanbaalenii PYR-1 | Mvan_0612 | - | 0.0 | 81.52% (460) | glutamate--ammonia ligase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3294|M.marinum_M -----------MDRQKEFVLRTLE----ERD----IRFVRLWFTDVLGFL
MUL_1335|M.ulcerans_Agy99 -----------MDRQKEFVLRTLE----ERD----IRFVRLWFTDVLGFL
MLBr_01631|M.leprae_Br4923 -----------MDRQKEFVLRTLE----ERD----IRFVRLWFTDVLGYL
TH_3396|M.thermoresistible__bu -----------MDRQKEFVLRSLE----ERD----IRFVRLWFTDVLGFL
MAB_1920|M.abscessus_ATCC_1997 -----------MDRQKEFVLRTLE----ERD----IRFVRLWFTDVLGYL
MSMEG_3828|M.smegmatis_MC2_155 ----------MSSTSALLDLDQFEKLAAAGE----ITTVICAAPDPYGRL
Mvan_0612|M.vanbaalenii_PYR-1 ----------MNDESTRLDLDQFEKLAAAGE----INTVVCATPDPYGRL
Mb2885c|M.bovis_AF2122/97 ---------MTGPGSPPLAWTELERLVAAGD----VDTVIVAFTDMQGRL
Rv2860c|M.tuberculosis_H37Rv ---------MTGPGSPPLAWTELERLVAAGD----VDTVIVAFTDMQGRL
MAV_3718|M.avium_104 --MASEDSALTGSDTAMLSLAALDRLVAAGAPETRVDTVIVAFPDMQGRL
Mflv_4065|M.gilvum_PYR-GCK MTDRTKGRLELMTTRGMLTLEDLEKQVADGD----IDTVIVAFCDMQGRL
: :: : * * * *
MMAR_3294|M.marinum_M KSVAIAPAELEGAFEEGIGFDGSSIEGFARVSE---------------SD
MUL_1335|M.ulcerans_Agy99 KSVAIAPAELEGAFEEGIGFDGSSIEGFARVSE---------------SD
MLBr_01631|M.leprae_Br4923 KSVAIAPAELEGAFEEGIGFDGSSIEGFARVSE---------------SD
TH_3396|M.thermoresistible__bu KSVAIAPAELEGAFEEGIGFDGSAIEGFARVSE---------------AD
MAB_1920|M.abscessus_ATCC_1997 KSVSIAPAELEGAFEEGIGFDGSSIEGFARVSE---------------SD
MSMEG_3828|M.smegmatis_MC2_155 VGKRLTISAFRTLGLDGGGVNASSFIFAVDLEMNPLD-LPVSNAANGWVD
Mvan_0612|M.vanbaalenii_PYR-1 VGKRLTVPAFRSLGLSGAGINASSFIFAVDLEMNPLD-LPVSNAANGWAD
Mb2885c|M.bovis_AF2122/97 AGKRISGRHFVDD-IATRGVECCSYLLAVDVDLNTVPGYAMASWDTGYGD
Rv2860c|M.tuberculosis_H37Rv AGKRISGRHFVDD-IATRGVECCSYLLAVDVDLNTVPGYAMASWDTGYGD
MAV_3718|M.avium_104 VGKRMDARLFVDE-AAATGVECCGYLLAVDVDMNTVGGYAISGWDTGFGD
Mflv_4065|M.gilvum_PYR-GCK TGKRISSRLFVDD-VAEHGAECCNYLLAVDVDMNTVDGYAISSWEKGYGD
. : : * : . . :. *
MMAR_3294|M.marinum_M TVAHPDPSTFQVLPWVTDAGHHHSARIFCDITMPDG-SPSWADPRHVLRR
MUL_1335|M.ulcerans_Agy99 TVAHPDPSTFQVLPWVTDAGHHHSARIFCDITMPDG-SPSWADPRHVLRR
MLBr_01631|M.leprae_Br4923 TVAHPDPSTFQILPWATTAGHHHSARMFCDITMPDG-SSSWADPRHVLRR
TH_3396|M.thermoresistible__bu MVARPDPSTFQVLPWTGDSGKHYSARMFCDITMPDG-SPSWADPRHVLRR
MAB_1920|M.abscessus_ATCC_1997 TVAHPDPSTFQVLPWTTSAGKHHSARMFCDIKLPDG-SPSWSDPRHVLRR
MSMEG_3828|M.smegmatis_MC2_155 IRLVPDLSTLRRVPWEPT-----AALVICDAYHMDSDEPLAVAPRTILRQ
Mvan_0612|M.vanbaalenii_PYR-1 IRLIPDLTTLRRVPWEPN-----VALVICDAYESHSDTMLSVAPRTILRR
Mb2885c|M.bovis_AF2122/97 MVMTPDLSTLRLIPWLPG-----TALVIADLVWADG-SEVAVSPRSILRR
Rv2860c|M.tuberculosis_H37Rv MVMTPDLSTLRLIPWLPG-----TALVIADLVWADG-SEVAVSPRSILRR
MAV_3718|M.avium_104 LVMRPDLSTLRRIPWLPG-----TALVIADVVGADG-SPVAVSPRAVLRR
Mflv_4065|M.gilvum_PYR-GCK MVMTPDMSTLRRVPWLPA-----TALVMADLSWTDG-RPVTQAPRGILRA
** :*:: :** * ::.* .. ** :**
MMAR_3294|M.marinum_M QLTKANELGFSCYVHPEIEFFLLNPGP----EDG--TEPVPIDNAGYFDQ
MUL_1335|M.ulcerans_Agy99 QLIKANELGFSCYVHPEIEFFLLNPGP----EDG--TEPVPIDNAGYFDQ
MLBr_01631|M.leprae_Br4923 QLTKANDLGFSCYVHPEIEFFLLKAGP----EDGSMPIPVPVDNAGYFDQ
TH_3396|M.thermoresistible__bu QLAKAGDQGFTCYVHPEIEFFLLKPGP----HDG--SPPVPADTSGYFDQ
MAB_1920|M.abscessus_ATCC_1997 QLAKASDLGFSCYVHPEIEFFLLKNLP----DDG--TAPIPADDAGYFDQ
MSMEG_3828|M.smegmatis_MC2_155 QIDRAAQQGLRFKFASELEFYLSCTPPRQAWDLGYQNLKMMSDYRSDYQM
Mvan_0612|M.vanbaalenii_PYR-1 QIERARQQGLTFKFASELEFFLSATPPRQAWELGYQDLKMLSDYRSDYQM
Mb2885c|M.bovis_AF2122/97 QLDRLKARGLVADVATELEFIVFDQPYRQAWASGYRGLTPASDYNIDYAI
Rv2860c|M.tuberculosis_H37Rv QLDRLKARGLVADVATELEFIVFDQPYRQAWASGYRGLTPASDYNIDYAI
MAV_3718|M.avium_104 QLDRLAGRGLFADAATELEFMVFDEPYRQAWASGYRGLTPASDYNIDYAI
Mflv_4065|M.gilvum_PYR-GCK QLDRLDALGLDAVAATELEFMVFDTGFRDAWAAGYRNLKPATDYNIDYAM
*: : *: .*:** : * * :
MMAR_3294|M.marinum_M AVHVSASKFRRHAIDALEFMGISVEFSHHEGAPGQQEIDLRFADALSMAD
MUL_1335|M.ulcerans_Agy99 AVHVSASKFRRHVIDALEFMGISVEFSHHEGAPGQQEIDLRFADALSMAD
MLBr_01631|M.leprae_Br4923 AVHDSALNFRRHAIEALEFMGISVEFSHHEGAPGQQEIDLRFADALSMAD
TH_3396|M.thermoresistible__bu SVHDTAPNFRRHAIEALEQMGISVEFSHHEGAPGQQEIDLRYADALSMAD
MAB_1920|M.abscessus_ATCC_1997 AVHEAAPNFRRHAIDALESMGISVEFSHHEGAPGQQEIDLRYADALSMAD
MSMEG_3828|M.smegmatis_MC2_155 IQAARDDWFIERIRNQMPEFGVPIESSKPEWGLGQQEVTLDYCDTLEMAD
Mvan_0612|M.vanbaalenii_PYR-1 IQSSRDDWFIERIRNQMPGFGVSIESSKPEWGLGQQEVTLDYCDTLEMAD
Mb2885c|M.bovis_AF2122/97 LASSRMEPLLRDIRLGMAGAGLRFEAVKGECNMGQQEIGFRYDEALVTCD
Rv2860c|M.tuberculosis_H37Rv LASSRMEPLLRDIRLGMAGAGLRFEAVKGECNMGQQEIGFRYDEALVTCD
MAV_3718|M.avium_104 SASSRMEPLLRDIRRGMAGAGLRFESVKGECNRGQQEIGFRYDEALRTCD
Mflv_4065|M.gilvum_PYR-GCK HASTRMEPLLRDIRLGMDGAGMYCEGVKGECNLGQQEIGFRYEQALVTCD
: . : *: * : * ****: : : ::* .*
MMAR_3294|M.marinum_M NVMTFRYVIKEVALEEGVRATFMPKPFGQHAGSAMHTHMSLFEGDVN---
MUL_1335|M.ulcerans_Agy99 NVMTFRYVIKEVALEEGVRATFMPKPFGQHAGSAMHTHMSLFEGDVN---
MLBr_01631|M.leprae_Br4923 NVMTFRYVIKEVAIENGARASFMPKPFAQHPGSAMHTHMSLFEGDVN---
TH_3396|M.thermoresistible__bu NVMTFRYVVKQVAISDGVRASFMPKPFAEHPGSAMHTHMSLFEGDTN---
MAB_1920|M.abscessus_ATCC_1997 NVMTFRNVVKEVAIIDGVHATFMPKPFSDHPGSAMHTHMSLFEGDTN---
MSMEG_3828|M.smegmatis_MC2_155 RHVLFKYGVKQLAHSADLTVTFMAKPKIDEVGSSCHLHVSMWSTDDDQ--
Mvan_0612|M.vanbaalenii_PYR-1 RHVLFKYGVKQLAHAADLTVTFMAKPKIDDVGSSCHLHVSMWSTDGE---
Mb2885c|M.bovis_AF2122/97 NHAIYKNGAKEIADQHGKSLTFMAKYDERE-GNSCHIHVSLRGTDGS---
Rv2860c|M.tuberculosis_H37Rv NHAIYKNGAKEIADQHGKSLTFMAKYDERE-GNSCHIHVSLRGTDGS---
MAV_3718|M.avium_104 NHVIYKNGAKEIADQHGKSLTFMAKYDERE-GNSCHVHLSLRDAQGG---
Mflv_4065|M.gilvum_PYR-GCK NHTIYKNGAKEIADQHGKSLTFMAKFDESE-GNSCHIHISFRGRDGSPED
. :: *::* . :**.* . *.: * *:*: :
MMAR_3294|M.marinum_M --AFHSPDDPLQLSEVGKSFIAGILEHASEISAVTNQWVNSYKRLILGGE
MUL_1335|M.ulcerans_Agy99 --AFHSPDDPLQLSEVGKSFIAGILEHASEISAVTNQWVNSYKRLILGGE
MLBr_01631|M.leprae_Br4923 --AFHSDDDPLQLSDVGKSFIAGILEHASEISAVTNQWVNSYKRLVVGGE
TH_3396|M.thermoresistible__bu --AFHSPDDPLQLSDVAKSFIAGILEHANEISAVTNQWVNSYKRLVHGGE
MAB_1920|M.abscessus_ATCC_1997 --AFHNPDDPLQLSDVGKSFIAGILEHANEISAVTNQWVNSYKRLVQGGE
MSMEG_3828|M.smegmatis_MC2_155 -PLCWCPE-RRGMSDRFGAFVAGQLRHAIDMSLMYAPTVNSYKRFKPDQF
Mvan_0612|M.vanbaalenii_PYR-1 -PLSWSTEPGGGMSATFGSFVAGQLTHATDIALLFAPTVNSYKRFQPDQF
Mb2885c|M.bovis_AF2122/97 -AVFADSNGPHGMSSMFRSFVAGQLATLREFTLCYAPTINSYKRFADSSF
Rv2860c|M.tuberculosis_H37Rv -AVFADSNGPHGMSSMFRSFVAGQLATLREFTLCYAPTINSYKRFADSSF
MAV_3718|M.avium_104 -AAFADPSRPHGMSTMFCSFLAGLLATMADFTLFYAPNINSYKRFADESF
Mflv_4065|M.gilvum_PYR-GCK IAVFADDSDELGMSSMFRSFIAGQLATLREFTLFYAPNINSYKRFAEGSF
. :* :*:** * ::: :*****:
MMAR_3294|M.marinum_M APTAASWGAANRSALVRVPMYTPHKTSSRRIEVRSPDSACNPYLAFAVLL
MUL_1335|M.ulcerans_Agy99 APTAASWGAANRSALVRVPMYTPHKTSSRRIEVRSPDSACNPYLAFAVLL
MLBr_01631|M.leprae_Br4923 APTTASWGAANRSALVRVPMYTPHKTSSRRVEVRSPDSACNPYLAFAVLL
TH_3396|M.thermoresistible__bu APTAASWGAANRSALVRVPMYTPHKASSRRVEVRSPDSACNPYLTFAVLL
MAB_1920|M.abscessus_ATCC_1997 APTAASWGAANRSALVRVPMYTPNKSSSRRIEVRSPDSACNPYLTYAVLL
MSMEG_3828|M.smegmatis_MC2_155 AGTAIALGMDNRSCAFRLVGNGP----SFRVENRIPGADVNPYYAYAATI
Mvan_0612|M.vanbaalenii_PYR-1 AGTAIALGNDNRSCAFRLVGGGP----SFRVENRIPGGDVNPYYAYSATI
Mb2885c|M.bovis_AF2122/97 APTALAWGLDNRTCALRVVGHGQ----NIRVECRVPGGDVNQYLAVAALI
Rv2860c|M.tuberculosis_H37Rv APTALAWGLDNRTCALRVVGHGQ----NIRVECRVPGGDVNQYLAVAALI
MAV_3718|M.avium_104 APTALAWGLDNRTCALRVVGHGA----HTRVECRVPGGDVNPYLAVAAIV
Mflv_4065|M.gilvum_PYR-GCK APTAVAWGLDNRTCSLRVVGHGQ----GMRMENRAPGGDVNQYLAVAALI
* *: : * **:. .*: *:* * *.. * * : :. :
MMAR_3294|M.marinum_M AAGLRGVEKGYVLGPQAEDNVWDLTPEERRTMGYRELPTSLDSALRAMES
MUL_1335|M.ulcerans_Agy99 AAGLRGVEKGYVLGPQAEDNVWDLTPEERRTMGYRELPTSLDSALRAMES
MLBr_01631|M.leprae_Br4923 AAGLRGVEKGYVLGPQAEDNVWDLTPEERLAMGYRELPTSLDSALGAMEV
TH_3396|M.thermoresistible__bu AAGLRGVEKGYVLGPEAEDNVWTLTPEERRAMGFKELPSSLGQALAEMER
MAB_1920|M.abscessus_ATCC_1997 AAGLRGVEQGYTLGPEAEDDVWGLTRAERQAMGYKELPSTLENALIAMEG
MSMEG_3828|M.smegmatis_MC2_155 AAGLDGVEANLPAPDIFDGNAWTREGVAT-------VPTSLHRSLDLFAE
Mvan_0612|M.vanbaalenii_PYR-1 AAGLAGIEHELPAPGIFDGNAWNGQDVPI-------VPTSMHEALRLFAE
Mb2885c|M.bovis_AF2122/97 AGGLYGIERGLQLPEPCVGNAYQGADVER-------LPVTLADAAVLFED
Rv2860c|M.tuberculosis_H37Rv AGGLYGIERGLQLPEPCVGNAYQGADVER-------LPVTLADAAVLFED
MAV_3718|M.avium_104 AGGLYGIEQGLALPEPCAGNAYRARGVGR-------LPATLAEAAALFEH
Mflv_4065|M.gilvum_PYR-GCK AGGLYGIENKLELEDAVEGNAYT-SGADR-------LPTTLSEAVELFEN
*.** *:* .:.: :* :: : :
MMAR_3294|M.marinum_M SELVAEALGEHVFDFFLRNKRREWATYR-SHVTPYELSTYLSL-
MUL_1335|M.ulcerans_Agy99 SELVAEALGEHVFDFFLRNKRREWATYR-SHVTPYELSTYLSL-
MLBr_01631|M.leprae_Br4923 SEFVAEALGEHVFDFFLRNKRAEWANYR-SHVSLYELKTYLSL-
TH_3396|M.thermoresistible__bu SELVAEALGEHVFDFFLRNKRAEWENYR-SHVTPYELKNYLSL-
MAB_1920|M.abscessus_ATCC_1997 SELVAEALGEHVFDFFLRNKRAEWARYR-SNVTPFELRAYLGL-
MSMEG_3828|M.smegmatis_MC2_155 SKMARTAFGDEVFDHLLSSAQGELEAFDSHTVTDWESRRYYERV
Mvan_0612|M.vanbaalenii_PYR-1 CKMARTAFGDEVFDHLLASSQSELVAFDSHTVTDWEKCRYYERV
Mb2885c|M.bovis_AF2122/97 SALVREAFGEDVVAHYLNNARVELAAFN-AAVTDWERIRGFERL
Rv2860c|M.tuberculosis_H37Rv SALVREAFGEDVVAHYLNNARVELAAFN-AAVTDWERIRGFERL
MAV_3718|M.avium_104 SALARQVFGDDVVAHYLNNARVELAAFH-AAVTDWERMRGFERL
Mflv_4065|M.gilvum_PYR-GCK SKIARDAFGDDVVDHYVNNARVELKAFN-AAVTDWERVRGFERL
. :. .:*:.*. . : . : * : *: :*