For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAAVRRFRRALPGDPEFGDPLSVAGDGGPRAAARVADRLLERDAASREVSLGALQVWQALTERVSGRPAN REVTLVFTDLVGFSSWSLRAGDDATLRLLRRVAQVAEPPLLEAGGHIVKRMGDGMMAVFGDPATAVRAVL VALDAVKGIEVDGYTPRMRVGIHTGRPQRIGSDWLGVDVNITARVMERAAKGGLMVSHATLAGIADGELA ELGVTVKRERRQLFSARPDGVPEDLVMYRVRTPSRLPAARPREQEPPDA*
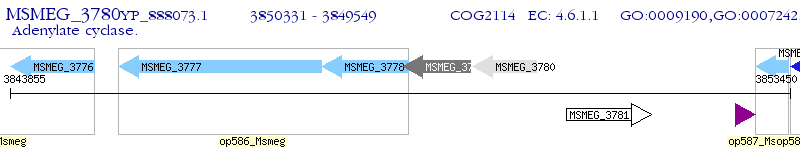
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3780 | - | - | 100% (260) | adenylate and guanylate cyclase catalytic domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_5018 | - | 5e-13 | 30.85% (188) | adenylate and guanylate cyclase catalytic domain-containing |
| M. smegmatis MC2 155 | MSMEG_4477 | - | 5e-12 | 34.13% (126) | hydrolase, alpha/beta hydrolase fold family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1674 | - | 1e-98 | 68.20% (261) | hypothetical protein Mb1674 |
| M. gilvum PYR-GCK | Mflv_3535 | - | 1e-107 | 77.37% (243) | putative adenylate/guanylate cyclase |
| M. tuberculosis H37Rv | Rv1647 | - | 1e-98 | 68.20% (261) | hypothetical protein Rv1647 |
| M. leprae Br4923 | MLBr_01399 | - | 2e-94 | 66.93% (251) | hypothetical protein MLBr_01399 |
| M. abscessus ATCC 19977 | MAB_2330 | - | 3e-91 | 66.40% (250) | putative purine cyclase-like protein |
| M. marinum M | MMAR_2454 | - | 1e-97 | 72.54% (244) | hypothetical protein MMAR_2454 |
| M. avium 104 | MAV_3122 | - | 1e-100 | 70.54% (258) | adenylate and guanylate cyclase catalytic domain-containing |
| M. thermoresistible (build 8) | TH_1590 | - | 1e-101 | 75.21% (242) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1633 | - | 9e-98 | 72.54% (244) | hypothetical protein MUL_1633 |
| M. vanbaalenii PYR-1 | Mvan_3321 | - | 1e-105 | 76.00% (250) | putative adenylate/guanylate cyclase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3535|M.gilvum_PYR-GCK -------------------------MPQSLSFRLVGVDEDPPGD----PG
Mvan_3321|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1590|M.thermoresistible__bu ---------------------------VELDGEVEPSDRRTRGP----DG
MSMEG_3780|M.smegmatis_MC2_155 --------------------------------------------------
Mb1674|M.bovis_AF2122/97 -------------------MPGSARTTYPCHVEVGPQDSESGAP---DET
Rv1647|M.tuberculosis_H37Rv -------------------MAGSARTTYPCHVEVGPQDSESGAP---DET
MMAR_2454|M.marinum_M -------------------MPGSATTTYPCEVEVGADNGDPGVP---DET
MUL_1633|M.ulcerans_Agy99 -------------------------------MEVGADNGDPGVP---DET
MLBr_01399|M.leprae_Br4923 -------------------MPPSAKSTYPGQVEGAPHDGTPSAPQEPDED
MAV_3122|M.avium_104 -------------------------------MELARRDLDPGEP---EES
MAB_2330|M.abscessus_ATCC_1997 MDVEPSGLDAEPDGEFDSGIRKRRRGRTRGPVRHDSAQVTDGIDTTADEV
Mflv_3535|M.gilvum_PYR-GCK G----GQFAGPRG---------WFRDINHHDGVIAFLRRTRRALPGDPDF
Mvan_3321|M.vanbaalenii_PYR-1 -----------MG---------WLMGVNHHDGVVAFLRRARRALPGDPDF
TH_1590|M.thermoresistible__bu AGFRSGRARGPLA---------WLKSANQNEGLVALVKRARRRLPGDPEF
MSMEG_3780|M.smegmatis_MC2_155 --------------------------------MVAAVRRFRRALPGDPEF
Mb1674|M.bovis_AF2122/97 ATAMASPVPRQRS------ALRWLRTVNRSPGLVSFIHRARRLLPGDPEF
Rv1647|M.tuberculosis_H37Rv ATAMASPVPRQRS------ALRWLRTVNRSPGLVSFIHRARRLLPGDPEF
MMAR_2454|M.marinum_M AITLAATAPRRRS------PVEWLRSTNHSPGVVAFVRRARRLLPGDPEF
MUL_1633|M.ulcerans_Agy99 AITLAATAPRRRS------PVEWLRSTNHSPGVVAFVRRARRLLPGDPEF
MLBr_01399|M.leprae_Br4923 TVTVPAPALIRRSSVSMPNAAQWLHTTNRSPRLVAMVRRARRLLPGDPDF
MAV_3122|M.avium_104 AAAQSASRRTALSVHN---ATQWLHNRNHSPGAVALVRRARRLLPGDPDF
MAB_2330|M.abscessus_ATCC_1997 PSTESASRHFARASR---TAGSWFRKMDRDPAVVAALRRARRSLPGDPYF
:: ::* ** ***** *
Mflv_3535|M.gilvum_PYR-GCK GDPLSVSGVGGPRAAARAADRLL-ERDAATREVSLGALQVWQALTERVSG
Mvan_3321|M.vanbaalenii_PYR-1 GDPLSVSGVGGPSAAARAADRYL-ERNAVTREVSLGALQVWQALTERVSG
TH_1590|M.thermoresistible__bu GDPLSVAGGDGPSVVARAAGRLL-ERDAATREVSLATLQVWQALTERISG
MSMEG_3780|M.smegmatis_MC2_155 GDPLSVAGDGGPRAAARVADRLL-ERDAASREVSLGALQVWQALTERVSG
Mb1674|M.bovis_AF2122/97 GDPLSTAGEGGPRAAARAADRLLRDRDAASREVGLSVLQVWQALTEAVSR
Rv1647|M.tuberculosis_H37Rv GDPLSTAGEGGPRAAARAADRLLRDRDAASREVGLSVLQVWQALTEAVSR
MMAR_2454|M.marinum_M GDPLSTAGDGGPRAAARAADRLLGDRDAASREVSLSVLQVWQAFTEAVSR
MUL_1633|M.ulcerans_Agy99 GDPLSTAGDGGPRAAARAADRLLGDRDAASREVSLSVLQVWQAFTEAVSR
MLBr_01399|M.leprae_Br4923 GDPLSTAGEGGPRAAARAADRLLGDRGAASREVSLSVLQVWQALTEAIAR
MAV_3122|M.avium_104 GDPLSTAGDGGPRAAARAADRLLGDRDAVSREVSLGVLQVWQALTEGISR
MAB_2330|M.abscessus_ATCC_1997 GDPLSTSGFGGASAVARAADRLVTDRDTAVREFGFGALQIWQAFTEKVSK
*****.:* .*. ..**.*.* : :*.:. **..:..**:***:** ::
Mflv_3535|M.gilvum_PYR-GCK RPANPEVTLVFTDLVGFSAWSLRSGDDATLRLLRRVAQVVEPPLLQSGGH
Mvan_3321|M.vanbaalenii_PYR-1 RPANAEVTLVFTDLVGFSTWSLGAGDDATLRLLRRVAQVVEPPLLEAGGH
TH_1590|M.thermoresistible__bu RPANPEVTLVFTDLVGFSSWALKAGDDATLRLLRRVAQVVEPPLLAAGGR
MSMEG_3780|M.smegmatis_MC2_155 RPANREVTLVFTDLVGFSSWSLRAGDDATLRLLRRVAQVAEPPLLEAGGH
Mb1674|M.bovis_AF2122/97 RPANPEVTLVFTDLVGFSTWSLHAGDDATLTLLRQVARAVESPLLDAGGH
Rv1647|M.tuberculosis_H37Rv RPANPEVTLVFTDLVGFSTWSLHAGDDATLTLLRQVARAVESPLLDAGGH
MMAR_2454|M.marinum_M RPANAEVTLVFTDLVGFSTWSLQAGDDAALKLLRQVARAVEPPLLDAGGH
MUL_1633|M.ulcerans_Agy99 RPANAEVTLVFTDLVGFSTWSLQAGDDAALKLLRQVARAVEPPLLDAGGH
MLBr_01399|M.leprae_Br4923 RPVNPEVTLVFTDLVGFSGWSLQAGDEATLALLRQVARAVESPLLDAGGH
MAV_3122|M.avium_104 RPANPEVTLVFTDLVGFSTWSLQAGDDAALALLRQVARAVEPPLLDAGGH
MAB_2330|M.abscessus_ATCC_1997 QPANREVTLVFTDLVGFSGWALEAGDEATLKLLRRVASVSEPPLLSAGGQ
:*.* ************* *:* :**:*:* ***:** . *.*** :**:
Mflv_3535|M.gilvum_PYR-GCK IVKRMGDGLMAVFADPTTAVRSAIAAREAVKTVDVDGYTPRMRVGIHTGR
Mvan_3321|M.vanbaalenii_PYR-1 IVKRMGDGLMAVFTDATTAVRAAIEARDAVKSLDVDGYTPRMRVGIHTGR
TH_1590|M.thermoresistible__bu IVKRMGDGSMVVFTEPVPAVRAALAAKDAVKSVEVDGYTPRMRVGVHTGR
MSMEG_3780|M.smegmatis_MC2_155 IVKRMGDGMMAVFGDPATAVRAVLVALDAVKGIEVDGYTPRMRVGIHTGR
Mb1674|M.bovis_AF2122/97 IVKRLGDGIMAVFRNPTVALRAVLVAQDAVKSLEVQGYTPRMRIGIHTGR
Rv1647|M.tuberculosis_H37Rv IVKRLGDGIMAVFRNPTVALRAVLVAQDAVKSLEVQGYTPRMRIGIHTGR
MMAR_2454|M.marinum_M IVKRLGDGIMAVFRNPTVAVRAVLIAQDAVKAVEVEGYTPRMRVGIHTGR
MUL_1633|M.ulcerans_Agy99 IVKRLGDGIMAVFRNPTVAVRAVLIAQDAVKAVEVEGYTPRMRVGIHTGR
MLBr_01399|M.leprae_Br4923 IVKRMGDGIMAVFRDPSVAVQAVLAATEAMKSVEVGGYTPRIRVGIHTGR
MAV_3122|M.avium_104 IVKRMGDGLMAVFGDPTVAVRAVLAAKEALRSVEVAGYTPRMRVGIHTGR
MAB_2330|M.abscessus_ATCC_1997 VVKRMGDGIMAVFDDPVTALRAIIPARDALKNVEIQGYTPVMRIGIHAGT
:***:*** *.** :. *::: : * :*:: ::: **** :*:*:*:*
Mflv_3535|M.gilvum_PYR-GCK PQRIGSDWLGIDVNLAARVMERATRGELVISEATLQRIPDEDFDVLPVTA
Mvan_3321|M.vanbaalenii_PYR-1 PQRIGSDWLGIDVNLAARVMERATRGELVVSGATLERIPDDEFQTLGVTA
TH_1590|M.thermoresistible__bu PQHIGSDWLGVDVNIAARVMERATRGGVMVSQTTLDRISDEDFDALNVTA
MSMEG_3780|M.smegmatis_MC2_155 PQRIGSDWLGVDVNITARVMERAAKGGLMVSHATLAGIADGELAELGVTV
Mb1674|M.bovis_AF2122/97 PQRLAADWLGVDVNIAARVMERATKGGIMISQPTLDLIPQSELDALGVVA
Rv1647|M.tuberculosis_H37Rv PQRLAADWLGVDVNIAARVMERATKGGIMISQPTLDLIPQSELDALGVVA
MMAR_2454|M.marinum_M PQRLAADWLGVDVNIAARVMERATKGGVMISSPTLDLIPQSELDAMRVVA
MUL_1633|M.ulcerans_Agy99 PQRLAADWLGVDVNIAARVMERATKGGVMISSPTLDLIPQSELDAMRVVA
MLBr_01399|M.leprae_Br4923 PQRLAADWLGVDVNIAARVMERATKGGIMISGPTLDLIPQSDLKELGIIT
MAV_3122|M.avium_104 PQRLASDWLGVDVNIAARVMERATKGGIMVSSSTLDHIPQSELDALGVEA
MAB_2330|M.abscessus_ATCC_1997 PQRIGSDWLGVDVNIAARVMESAAKGGLAISQPALAKVPLAFLDHMGLTP
**::.:****:***::***** *::* : :* .:* :. : : :
Mflv_3535|M.gilvum_PYR-GCK KKLRRQVFAARADGVPDDFTMYRLRTRRPVRGHEDDHDEVPGEE--
Mvan_3321|M.vanbaalenii_PYR-1 KKLRRPVFAGKQDGVPGDFAMYRLRTHRQSPGAEEG--VVAGG---
TH_1590|M.thermoresistible__bu KRVRRSLFAPQPDGVPEGLTVYRLRAAAGGTGPSGG-PAESGEKPR
MSMEG_3780|M.smegmatis_MC2_155 KRERRQLFSARPDGVPEDLVMYRVRTPSRLPAARPREQEPPDA---
Mb1674|M.bovis_AF2122/97 RRVRKPVFASKPTGIPPDLAIYRIKTVSESTAADNFDEMSPDAQ--
Rv1647|M.tuberculosis_H37Rv RRVRKPVFASKPTGIPPDLAIYRIKTVSESTAADNFDEMSPDAQ--
MMAR_2454|M.marinum_M KRARRPMFASKLTGIPPDLAIYRLTTRKDLATIGDTADTDLQA---
MUL_1633|M.ulcerans_Agy99 KRARRPMFASKLTGIPPDLAIYRLTTRKDLATIGDTADTDLQA---
MLBr_01399|M.leprae_Br4923 RRVRKPMFTSKFTGIPPDMVIYRIKARRELTASDETAQTNSLT---
MAV_3122|M.avium_104 KRTRKPVFGPKPAGMPADLAIYRLKTLKELSASDDTDETKPQP---
MAB_2330|M.abscessus_ATCC_1997 TPVRRPLFSSRPAGVPENMKMYLLETRRQLPAPGDDGRV-------
*: :* : *:* .: :* : :