For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NEIPDVVNAVDVEAWRDEAADEVDVVVIGFGIAGGCAAVSAAAAGARVLVLERAAAAGGTSAMAGGHFYL GGGTAVQRATGHDDTPDEMYKYLVAVAKDPDHAKIRAYCDGSVEHFNWLEALGFEFERTYYPGKVVVPPG TEGLSYTGNEKVWPYREQARPAPRGHSVPVPGELGGAAMVIDLLLKRAEELGVEIRYEHGVTNLVVDDGA VAGVKWKHFGETGCVKARSVIIAAGGFAMNPEMVAQYTPALGAKRKTKHHGEVEPYILGNPNDDGLGIRL GVSAGGVARDMDQLFITAAAYPPEILLTGIIVNNRGERFVAEDSYHSRTSAFVLEQPDQEAYLIVDEAHM EMPAMPLIKFIDGYETVEEMATALGIPVENLTATLQRYNENARRGEDPDFHKDPAYVAPQDSGPWAAFDL SLGRAMYSGFTMGGLTVSIDGEVLREDGSAIPGLYAAGACASNIAQDGKGYASGTQLGEGSFFGRRAGAH AASRVRVG*
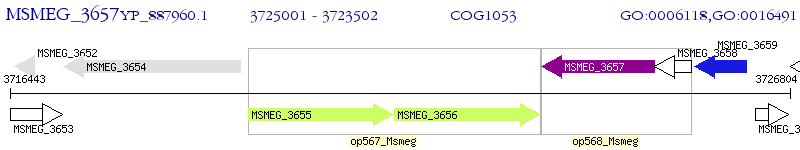
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3657 | - | - | 100% (499) | hypothetical protein MSMEG_3657 |
| M. smegmatis MC2 155 | MSMEG_2873 | - | 2e-59 | 31.61% (541) | hypothetical protein MSMEG_2873 |
| M. smegmatis MC2 155 | MSMEG_4864 | - | 5e-36 | 26.13% (574) | 3-ketosteroid-delta-1-dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1848 | - | 0.0 | 71.77% (496) | hypothetical protein Mb1848 |
| M. gilvum PYR-GCK | Mflv_3411 | - | 0.0 | 81.65% (496) | hypothetical protein Mflv_3411 |
| M. tuberculosis H37Rv | Rv1817 | - | 0.0 | 71.77% (496) | hypothetical protein Rv1817 |
| M. leprae Br4923 | MLBr_00697 | sdhA | 1e-08 | 42.19% (64) | succinate dehydrogenase flavoprotein subunit |
| M. abscessus ATCC 19977 | MAB_2389 | - | 0.0 | 68.35% (496) | hypothetical protein MAB_2389 |
| M. marinum M | MMAR_2694 | - | 0.0 | 73.79% (477) | flavoprotein |
| M. avium 104 | MAV_2897 | - | 0.0 | 79.76% (494) | hypothetical protein MAV_2897 |
| M. thermoresistible (build 8) | TH_4191 | - | 9e-61 | 32.10% (542) | PUTATIVE 3-ketosteroid-delta4 |
| M. ulcerans Agy99 | MUL_3063 | - | 0.0 | 71.37% (496) | flavoprotein |
| M. vanbaalenii PYR-1 | Mvan_3165 | - | 0.0 | 81.45% (496) | hypothetical protein Mvan_3165 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1848|M.bovis_AF2122/97 --------------------------------------------------
Rv1817|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2694|M.marinum_M --------------------------------------------------
MUL_3063|M.ulcerans_Agy99 MNLSVRSPTQIAWVTPDLDATEAAFTGLLGARKWVRIPDVHFAPDSCSYR
MAB_2389|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3411|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3165|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2897|M.avium_104 --------------------------------------------------
MSMEG_3657|M.smegmatis_MC2_155 --------------------------------------------------
TH_4191|M.thermoresistible__bu --------------------------------------------------
MLBr_00697|M.leprae_Br4923 --------------------------------------------------
Mb1848|M.bovis_AF2122/97 --------------------------------------------------
Rv1817|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2694|M.marinum_M --------------------------------------------------
MUL_3063|M.ulcerans_Agy99 GKPADFVASISLSYLGDMQLELISPVRGENIYSDFPRDCGPGLHHICLEV
MAB_2389|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3411|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3165|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2897|M.avium_104 --------------------------------------------------
MSMEG_3657|M.smegmatis_MC2_155 --------------------------------------------------
TH_4191|M.thermoresistible__bu --------------------------------------------------
MLBr_00697|M.leprae_Br4923 --------------------------------------------------
Mb1848|M.bovis_AF2122/97 --------------------------------------------------
Rv1817|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2694|M.marinum_M --------------------------------------------------
MUL_3063|M.ulcerans_Agy99 EGPEQLEIALAEAADHGASVVQRGSMPGGIQFAYVAAPQAGVPFVEIAYF
MAB_2389|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3411|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3165|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2897|M.avium_104 --------------------------------------------------
MSMEG_3657|M.smegmatis_MC2_155 --------------------------------------------------
TH_4191|M.thermoresistible__bu --------------------------------------------------
MLBr_00697|M.leprae_Br4923 --------------------------------------------------
Mb1848|M.bovis_AF2122/97 ---------------MSTDIPATVSAETVTSW----SDDVDVTVIGFGIA
Rv1817|M.tuberculosis_H37Rv ---------------MSTDIPATVSAETVTSW----SDDVDVTVIGFGIA
MMAR_2694|M.marinum_M ----------------------------MTSW----TDDVDVVVIGFGIA
MUL_3063|M.ulcerans_Agy99 SPEIKAFYDYIKRAALNTEIPATVSSDTVTSW----TDDVDVVVIGFGIA
MAB_2389|M.abscessus_ATCC_1997 ---------------MSAAIPETVAVQDITEW----SDEVDVLVIGFGMG
Mflv_3411|M.gilvum_PYR-GCK ----------------MQHIPDTVSVEDVTSW----SDETDVVVIGFGIA
Mvan_3165|M.vanbaalenii_PYR-1 ----------------MQHIPETVNVGDVTSW----SDEVDVVVVGFGIA
MAV_2897|M.avium_104 ---------------MSTEIPATVSAHDVTSW----SDDVDVVVIGFGIA
MSMEG_3657|M.smegmatis_MC2_155 ----------------MNEIPDVVNAVDVEAWRDEAADEVDVVVIGFGIA
TH_4191|M.thermoresistible__bu -------------------MPT--------------AQEYDVIIVGFGAA
MLBr_00697|M.leprae_Br4923 ---------MIQQHRYDVVIVGAGGAGMRAAVEAGPRVRTAVLTKLYPTR
* :
Mb1848|M.bovis_AF2122/97 GGCAAVSAAAAGARVLVLERAAAAGGTTALAGGHFYLGGGTTVQLATGHP
Rv1817|M.tuberculosis_H37Rv GGCAAVSAAAAGARVLVLERAAAAGGTTALAGGHFYLGGGTTVQLATGHP
MMAR_2694|M.marinum_M GGCAAVAAAAAGARVLVLERAAAAGGTTSLAGGHFYLGGGTAVQQATGHS
MUL_3063|M.ulcerans_Agy99 GGCAAVAAAAAGARVLVLERAAAAGGTTSLAGGHFYLGGGTAVQQATGHS
MAB_2389|M.abscessus_ATCC_1997 GGCAAVSAAEAGAQVLALERAASAGGTTAMAGGHFYLGGGTAVQQATGHA
Mflv_3411|M.gilvum_PYR-GCK GGSAAVSAAAAGARVLVLEKAAAAGGTTSMAGGHFYLGGGTAVQQATGHD
Mvan_3165|M.vanbaalenii_PYR-1 GGCAAVSAAAAGARVLVLEKAAAVGGTTSMAGGHFYLGGGTAVQQATGHD
MAV_2897|M.avium_104 GGCAAVSAAADGARVLVLERAAAAGGTTSLAGGHFYLGGGTAVQQATGHD
MSMEG_3657|M.smegmatis_MC2_155 GGCAAVSAAAAGARVLVLERAAAAGGTSAMAGGHFYLGGGTAVQRATGHD
TH_4191|M.thermoresistible__bu GASAAIEAADRGARVLVLDRGHGGG-ATAYSGGIVYAGAGTAEQQAGGHA
MLBr_00697|M.leprae_Br4923 SHTGAAQGGMCAALANVEDDNWEWHAFDTVKGGDYLADQDAVEIMCKEAI
. .* .. .* . . : : ** . .:. .
Mb1848|M.bovis_AF2122/97 DSPEEMYKYLVAVSREPD-HDKIRAYCDGSVEH-FNWLEGLGFQFER-SY
Rv1817|M.tuberculosis_H37Rv DSPEEMYKYLVAVSREPD-HDKIRAYCDGSVEH-FNWLEGLGFQFER-SY
MMAR_2694|M.marinum_M DTPDEMYKYLVAVSRQPD-HAKIRAYCDGSVEH-FNWLEDLGFQFER-SY
MUL_3063|M.ulcerans_Agy99 DTPDEMYKYLAAVSRQPD-HAKIRAYCDGSVEH-FNWLEDLGFQFER-SY
MAB_2389|M.abscessus_ATCC_1997 DSADEMYKYLMAVSPDPD-PEKIRLYCDGSVEH-FNWLEALGFQFER-SF
Mflv_3411|M.gilvum_PYR-GCK DSPEEMYKYLVTQSRNPE-HDKIRAYCDGSVEH-FNWLEALGFQFER-SY
Mvan_3165|M.vanbaalenii_PYR-1 DSAEEMYKYLVSQSRDPE-HDKIRAYCEGSVEH-FNWLEALGFQFER-SY
MAV_2897|M.avium_104 DSAEEMYKYLVAVSREPE-LDKIRAYCEGSVEH-FNWLEDLGFQFER-SY
MSMEG_3657|M.smegmatis_MC2_155 DTPDEMYKYLVAVAKDPD-HAKIRAYCDGSVEH-FNWLEALGFEFER-TY
TH_4191|M.thermoresistible__bu DSVENMRAYLAREVGDAVSPETLTRFCEQSPAM-IEWLKAQGVEFRGGPV
MLBr_00697|M.leprae_Br4923 DAVLDLEKMGMPFNRTPEGRIDQRRFGGHTRDHGKAPVRRACYAADRTGH
*: :: . : : :.
Mb1848|M.bovis_AF2122/97 FPGKAVIQ--PNTEGLMFTGNEKVWPFLELAVPAPRGHKVPVPGDTGGAA
Rv1817|M.tuberculosis_H37Rv FPGKAVIQ--PNTEGLMFTGNEKVWPFLELAVPAPRGHKVPVPGDTGGAA
MMAR_2694|M.marinum_M FPGKAVIQ--PNTEGLMFTGNEKVWPFFEKAVPAPRGHKVPVPGDTGGAS
MUL_3063|M.ulcerans_Agy99 FPGKAVIQ--PNTEGLMFTGNEKVWPFFEKAVPAPRGHKVPVPGDTGGAS
MAB_2389|M.abscessus_ATCC_1997 YPEKAVIQ--PNTEGLMFTGNELVWPYRDEAIPAPRGHKVPKPGDTGGAG
Mflv_3411|M.gilvum_PYR-GCK YPGKVVVP--PGTEGLSYTGNEKVWPFCEQARPAPRGHSVPVPGELGGAA
Mvan_3165|M.vanbaalenii_PYR-1 YPGKVVVP--PGTEGLSYTGNEKVWPFCEQAKPAPRGHSVPVPGELGGAA
MAV_2897|M.avium_104 YPGKVVVP--PGTEGLSYTGNEKVWPFCEQAKPAPRGHSVPVPGELGGAA
MSMEG_3657|M.smegmatis_MC2_155 YPGKVVVP--PGTEGLSYTGNEKVWPYREQARPAPRGHSVPVPGELGGAA
TH_4191|M.thermoresistible__bu PPYKTSYP--TDEHYLYYSGNETAFPYSEHAHPAPRGHRALAPG-MRSGR
MLBr_00697|M.leprae_Br4923 MILQTLYQNCVKHDVEFFNEFYALDLVLTQTPSGPVTTGVVAYELATGDI
:. . :. : ..* . .
Mb1848|M.bovis_AF2122/97 MVIDLLLKRAASLGIQIRYETGATELIVDGTG---------KVTGVMWKR
Rv1817|M.tuberculosis_H37Rv MVIDLLLKRAASLGIQIRYETGATELIVDGTG---------KVTGVMWKR
MMAR_2694|M.marinum_M MVIDLLLKRAASLGVQIRYETGATELIVDGSGT------AARVAGVMWKR
MUL_3063|M.ulcerans_Agy99 MVIDLLLKRGASLGVQIRYETGATELIVAGSGT------AARVAGVMWKR
MAB_2389|M.abscessus_ATCC_1997 MVVELLAQRAAELGIPIRYETGATALILDEGG---------AVVGVTWKH
Mflv_3411|M.gilvum_PYR-GCK MVIDLLVKRADELGVQMRYETGATALITDADG---------AVVGVRWKH
Mvan_3165|M.vanbaalenii_PYR-1 MVIDLLLKRAADLGVQIRYETGVTGLVVDDDG---------AVVGVRWKH
MAV_2897|M.avium_104 MVIDLLVKRAEALGVQIRYETGATNLIVDDDG---------AVVGVAWKH
MSMEG_3657|M.smegmatis_MC2_155 MVIDLLLKRAEELGVEIRYEHGVTNLVVD-DG---------AVAGVKWKH
TH_4191|M.thermoresistible__bu VLFERLRDSALSKGVEFVPLAHVHSLIQADDG---------RVTGVRYTV
MLBr_00697|M.leprae_Br4923 HVFHTKAVVIATGGSGRMYKTTSNAHTLTGDGIGIVFRKGLPLEDMEFHQ
:.. * * : .: :
Mb1848|M.bovis_AF2122/97 FSETG------------------------------------------AIK
Rv1817|M.tuberculosis_H37Rv FSETG------------------------------------------AIK
MMAR_2694|M.marinum_M FTETG------------------------------------------AIK
MUL_3063|M.ulcerans_Agy99 FTETG------------------------------------------AIK
MAB_2389|M.abscessus_ATCC_1997 FTETG------------------------------------------AIR
Mflv_3411|M.gilvum_PYR-GCK FTETG------------------------------------------DIK
Mvan_3165|M.vanbaalenii_PYR-1 FTETG------------------------------------------EVK
MAV_2897|M.avium_104 FTETG------------------------------------------AVK
MSMEG_3657|M.smegmatis_MC2_155 FGETG------------------------------------------CVK
TH_4191|M.thermoresistible__bu LDPGHRHARRHRLITRATGKLGTWMPGVVSGAVRYAERLRAEASVETETR
MLBr_00697|M.leprae_Br4923 FHPTGLAG--------------------------------------LGIL
:
Mb1848|M.bovis_AF2122/97 AKSVIIAAGGFVMNPD---MVAKYTPKLAEK----------PFVLGNTYD
Rv1817|M.tuberculosis_H37Rv AKSVIIAAGGFVMNPD---MVAKYTPKLAEK----------PFVLGNTYD
MMAR_2694|M.marinum_M AKSVIIAAGGFVMNPE---MVAKYTPKLAEK----------PFVLGNTYD
MUL_3063|M.ulcerans_Agy99 AKSVIIAAGGFVMNPE---MVAKYTPKLAEK----------PFVLGNTYD
MAB_2389|M.abscessus_ATCC_1997 AKSVIIAAGGFVMNPA---MVAEYTPKLAEK----------PFVLGSTYD
Mflv_3411|M.gilvum_PYR-GCK ANSVIIAAGGFAMNPE---MVAEYTPALGQERKTKHHGNVAPYILGNPND
Mvan_3165|M.vanbaalenii_PYR-1 AKSVVIAAGGFAMNAE---MVAEYTPALAAERKTKHHGTVAPYILGNPND
MAV_2897|M.avium_104 ADAVILAAGGFAMNPQ---MVAEHTPALGQPRRTKHHGLVAPYILGNPHD
MSMEG_3657|M.smegmatis_MC2_155 ARSVIIAAGGFAMNPE---MVAQYTPALGAKRKTKHHGEVEPYILGNPND
TH_4191|M.thermoresistible__bu AGAVILAAGGFINNRG---WVRQYAPQFLKIS-----------PLGTVGD
MLBr_00697|M.leprae_Br4923 ISEAVRGEGGRLLNGENERFMEHYAPTIVDLAPRDIVAR--SMVLEVLEG
.: . ** * : .::* : * .
Mb1848|M.bovis_AF2122/97 DGLGIRLG---------------------VSAGGATQHMDQMFITAPPYP
Rv1817|M.tuberculosis_H37Rv DGLGIRLG---------------------VSAGGATQHMDQMFITAPPYP
MMAR_2694|M.marinum_M DGLGIRMG---------------------VSAGGATEHMDQIFVTAPPYP
MUL_3063|M.ulcerans_Agy99 DGLGIRMG---------------------VSAGGATEHMDQIFVTAPPYP
MAB_2389|M.abscessus_ATCC_1997 DGLGIRLG---------------------VSAGGATRNMNQIFVTAPVYP
Mflv_3411|M.gilvum_PYR-GCK DGLGIKLG---------------------VSAGGATDNLDQLFITAAAYP
Mvan_3165|M.vanbaalenii_PYR-1 DGLGIKLG---------------------VSAGGVAANLDQLFITAAAYP
MAV_2897|M.avium_104 DGLAIRMG---------------------VSAGGATKHMDELFITAAAYP
MSMEG_3657|M.smegmatis_MC2_155 DGLGIRLG---------------------VSAGGVARDMDQLFITAAAYP
TH_4191|M.thermoresistible__bu DGTGIRLG---------------------LDAGGKTDKMGNVTAWRFLSP
MLBr_00697|M.leprae_Br4923 RGAGPHKDYVYIDVRHLGEEVLESKLPDITEFSRTYLGVDPVHELVPVYP
* . : . . . :. : *
Mb1848|M.bovis_AF2122/97 PSILLTGIIVNKLGQRFVAEDS------YHSRTAGFIMEQPD---SAAYL
Rv1817|M.tuberculosis_H37Rv PSILLTGIIVNKLGQRFVAEDS------YHSRTAGFIMEQPD---SAAYL
MMAR_2694|M.marinum_M PSILLTGVIVNKLGRRFVAEDS------YHSRTAGFIMDQPD---SAAYL
MUL_3063|M.ulcerans_Agy99 PSILLTGVIVNKLGRRFVAEDS------YHSRTAGFIMDQPD---SAAYL
MAB_2389|M.abscessus_ATCC_1997 PSGLLTGIIVNKNGERFVSEDS------YHSRTSGFVMDQVD---SAAYL
Mflv_3411|M.gilvum_PYR-GCK PEILLTGIIVNKNGDRFVAEDS------YHSRTSAFVLEQPD---QTAYL
Mvan_3165|M.vanbaalenii_PYR-1 PEILLTGVIVNKDGQRFVAEDS------YHSRTSAFVLEQPE---QTAYL
MAV_2897|M.avium_104 PEVLLTGIIVNKEGKRFVAEDS------YHSRTSAFVLEQPE---QTAYL
MSMEG_3657|M.smegmatis_MC2_155 PEILLTGIIVNNRGERFVAEDS------YHSRTSAFVLEQPD---QEAYL
TH_4191|M.thermoresistible__bu PSAFLEGLTVGADGRRIANEDL------YGATHGDVLMREFG---GTGWA
MLBr_00697|M.leprae_Br4923 TCHYVMGGIPTTVTGQVLRDNTSTVPGLYAAGECACVSVHGANRLGTNSL
. : * :. :: * : : .
Mb1848|M.bovis_AF2122/97 IVDEAHLEHPKMPLVPLIDGWE----------------TVVEMEAALGIP
Rv1817|M.tuberculosis_H37Rv IVDEAHLEHPKMPLVPLIDGWE----------------TVVEMEAALGIP
MMAR_2694|M.marinum_M IVDEAHLQHPKMPLVPLIDGWE----------------SVAEMEAALGIP
MUL_3063|M.ulcerans_Agy99 IVDEAHLQHPKMPLVPLIDGWE----------------SVAEMEAALGIP
MAB_2389|M.abscessus_ATCC_1997 IVDSEHMDRPTLPLTPFIDGWE----------------TVAEMEAALGIP
Mflv_3411|M.gilvum_PYR-GCK IVDEAHMQMPEMPLIKFLDGYE----------------TIEELEKELGIP
Mvan_3165|M.vanbaalenii_PYR-1 IVDEDHMQMPEMPLIKFVDGFE----------------TIAEIESALGIP
MAV_2897|M.avium_104 IVDEAHTEMPEMPLIRFLDGYE----------------TIAEMEAALGIP
MSMEG_3657|M.smegmatis_MC2_155 IVDEAHMEMPAMPLIKFIDGYE----------------TVEEMATALGIP
TH_4191|M.thermoresistible__bu IYDAATWRKIRTQITDQTQVFQRLQVWYLLTIGHKKAATIADLARKNGID
MLBr_00697|M.leprae_Br4923 LDINVFGRRSGIAAASYASANDFVD------------MPPNPAEMVISWV
: . : . .
Mb1848|M.bovis_AF2122/97 PGNLAATLDRYNAYAARGADPDFHKQPEFLAAQDNGPWGAFDMSLGKAMY
Rv1817|M.tuberculosis_H37Rv PGNLAATLDRYNAYAARGADPDFHKQPEFLAAQDNGPWGAFDMSLGKAMY
MMAR_2694|M.marinum_M DGNLVATLDRYNTYAARGEDPDFHKQPEFLAPQDKGPWGAFDMSLGKAMY
MUL_3063|M.ulcerans_Agy99 DCNLVAALDRYNTYAARGEDPDFHKQPEFLAPQDKGPWGAFDMSLGKAMY
MAB_2389|M.abscessus_ATCC_1997 AGNLDKTLQRYNEYAAKGEDPDFHKHPKYLAAQGTGPWGAFDLTLGKAMY
Mflv_3411|M.gilvum_PYR-GCK AGRVAATLERYNTNAAAGEDPDFHKQSEYVAPQDKGPWAVFDLSLGRAMY
Mvan_3165|M.vanbaalenii_PYR-1 DGNLAATLECYNANAAAGVDPDFHKQPEYLAAQDKGPWAVFDLSLGRAMY
MAV_2897|M.avium_104 AGNLAATLERYNKFAAAGEDPDFHKQPEYLAAQDNGPWAVFDLSLGRAMY
MSMEG_3657|M.smegmatis_MC2_155 VENLTATLQRYNENARRGEDPDFHKDPAYVAPQDSGPWAAFDLSLGRAMY
TH_4191|M.thermoresistible__bu AAGLQQTVDEYNRGISSGAGDPGHKHPSLCPPLERGPFYSINISADASLF
MLBr_00697|M.leprae_Br4923 ANILSEHGNERVADIRGALQQTMDNNAAVFRTEETLKQALTDIHSLKERY
: : . .:.. . :: :
Mb1848|M.bovis_AF2122/97 A---GFTLG-----GLATSVD-GQVLRDDGAVVAGLYAVGACASNIAQDG
Rv1817|M.tuberculosis_H37Rv A---GFTLG-----GLATSVD-GQVLRDDGAVVAGLYAVGACASNIAQDG
MMAR_2694|M.marinum_M A---GFTLG-----GLATSVD-GEVLRSDGSVVAGLYAAGACASNIAQDG
MUL_3063|M.ulcerans_Agy99 A---GFTLG-----GLATSVD-GEVLRSDGSVVAGLYAAGACASNIAQDG
MAB_2389|M.abscessus_ATCC_1997 A---GFTLG-----GLRTDAD-GRVLTESGEAIPGLYAAGASASNIAQDG
Mflv_3411|M.gilvum_PYR-GCK S---GFTMG-----GLTVSID-GEVLRGDRSAIPGLYAAGACACNIAQDG
Mvan_3165|M.vanbaalenii_PYR-1 S---GFTMG-----GLTVSID-GEVLREDGSAIPGLYAAGACACNIAQDG
MAV_2897|M.avium_104 S---GFTIG-----GLAVSVD-GEVQRADGSVVPGLYAAGACASNLAQDG
MSMEG_3657|M.smegmatis_MC2_155 S---GFTMG-----GLTVSID-GEVLREDGSAIPGLYAAGACASNIAQDG
TH_4191|M.thermoresistible__bu YPIPGLTLG-----GLVVDEDSGAVVHRDGGVIPGLYAAGRNAVGVCSN-
MLBr_00697|M.leprae_Br4923 SRITVHDKGKSFNSDLLEAIELGFLLELAEVTVVGALNRKESRGGHARED
* .* : * : .: * . . :
Mb1848|M.bovis_AF2122/97 KGYASGTQLGEGSFFGRRAGAHAAARAQGM--------------------
Rv1817|M.tuberculosis_H37Rv KGYASGTQLGEGSFFGRRAGAHAAARAQGM--------------------
MMAR_2694|M.marinum_M KGYASGTQLGEGSFFGRRAGAHAAAHAGARVAP-----------------
MUL_3063|M.ulcerans_Agy99 KGYASGTQLGEGSFFGRRAGAHAAAHACARVAP-----------------
MAB_2389|M.abscessus_ATCC_1997 KGYSSGTRLGEASFFGRRAGSHASVRVSVA--------------------
Mflv_3411|M.gilvum_PYR-GCK KGYASGTQLGEGSFFGRRAGEHAARRAGEHAARR----------------
Mvan_3165|M.vanbaalenii_PYR-1 KGYASGTQLGEGSFFGRRAGEAAARRAGEAAARRAGEAAARRAGEAAARR
MAV_2897|M.avium_104 KGYASGTQLGEGSFFGRRAGAHAARRAAKA--------------------
MSMEG_3657|M.smegmatis_MC2_155 KGYASGTQLGEGSFFGRRAGAHAASRVRVG--------------------
TH_4191|M.thermoresistible__bu -SYVSGLSISDCVFSGRRAGAHAAGGTR----------------------
MLBr_00697|M.leprae_Br4923 YPHRDDTNYMRHTMAYKQGTDLLSDIALDFKPVVQTRYEPQERKY-----
: .. : ::. : .
Mb1848|M.bovis_AF2122/97 ---------
Rv1817|M.tuberculosis_H37Rv ---------
MMAR_2694|M.marinum_M ---------
MUL_3063|M.ulcerans_Agy99 ---------
MAB_2389|M.abscessus_ATCC_1997 ---------
Mflv_3411|M.gilvum_PYR-GCK ---------
Mvan_3165|M.vanbaalenii_PYR-1 AGEAAAERA
MAV_2897|M.avium_104 ---------
MSMEG_3657|M.smegmatis_MC2_155 ---------
TH_4191|M.thermoresistible__bu ---------
MLBr_00697|M.leprae_Br4923 ---------