For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NLQIDDSNRVRTLTLNRPDALNAFNEALYDEVTDALLAAGGDPEVSVVLLTGAGRAFSSGNDLVEMQTRI ANPDFTPGEHGFYGMIEALADFPKPFLCAVNGVGVGIGATILGFADLVFMADTARLKCPFTSLGVAPEAA SSYLMPRLIGRQNAAWLLMSSEWISAAEAKEMGLVFKVCTPGDLLTEARRHAEILAAKPVTSLQAVKETM VAPVREAVAAATRRESELFVELVGAAANAEALAAFTGRKGAAGS*
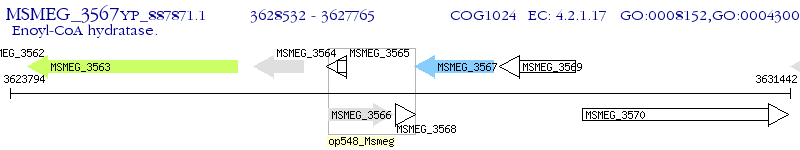
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3567 | - | - | 100% (255) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_0340 | - | 7e-83 | 61.29% (248) | enoyl-CoA hydratase/isomerase family protein |
| M. smegmatis MC2 155 | MSMEG_4299 | - | 2e-31 | 32.13% (249) | enoyl-CoA hydratase/isomerase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1099c | echA8 | 7e-31 | 31.30% (246) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_3325 | - | 1e-90 | 68.83% (247) | enoyl-CoA hydratase/isomerase |
| M. tuberculosis H37Rv | Rv1070c | echA8 | 7e-31 | 31.30% (246) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 3e-30 | 32.11% (246) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_1185c | - | 4e-27 | 29.51% (244) | enoyl-CoA hydratase |
| M. marinum M | MMAR_2759 | echA8_6 | 1e-100 | 71.20% (250) | enoyl-CoA hydratase, EchA8_6 |
| M. avium 104 | MAV_2835 | - | 1e-102 | 72.29% (249) | enoyl-CoA hydratase/isomerase family protein |
| M. thermoresistible (build 8) | TH_3154 | - | 1e-100 | 72.18% (248) | PUTATIVE Enoyl-CoA hydratase/isomerase |
| M. ulcerans Agy99 | MUL_2995 | echA8_6 | 2e-98 | 70.40% (250) | enoyl-CoA hydratase, EchA8_6 |
| M. vanbaalenii PYR-1 | Mvan_3046 | - | 1e-91 | 68.00% (250) | enoyl-CoA hydratase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3325|M.gilvum_PYR-GCK ------MTLQISDENRVRTLVLNRPDALNAFNQELYLATATALRDAADDP
Mvan_3046|M.vanbaalenii_PYR-1 ------MTLQITDENRVRTLVLNRPEALNAFNQELYLATATALREAADDP
MMAR_2759|M.marinum_M ------MTLQIDDDNRVRTLTLNRPEALNAFNEALYDATAQALLDAADDP
MUL_2995|M.ulcerans_Agy99 ------MTLQIDDDNRVRTLTLNRPEALNAFNEALYDATAQALWDAADDP
MAV_2835|M.avium_104 ------MTLQIDDDKRVRTLTLNRPEALNAFNEALYDATAEALLAAAEDP
TH_3154|M.thermoresistible__bu --------VQISDVNKVRTLTLDRPEALNAFNEALYDATTVALRDADADP
MSMEG_3567|M.smegmatis_MC2_155 ------MNLQIDDSNRVRTLTLNRPDALNAFNEALYDEVTDALLAAGGDP
Mb1099c|M.bovis_AF2122/97 MT---YETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAATELDDDP
Rv1070c|M.tuberculosis_H37Rv MT---YETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAATELDDDP
MLBr_02402|M.leprae_Br4923 MK---YDTILVDGYQRVGIITLNRPQALNALNSQMMNEITNAAKELDIDP
MAB_1185c|M.abscessus_ATCC_199 MTSHNFETILTERIDRVAVITLNRPKALNALNSQVMNEVTTAAAEFDADH
: .:* :.*:**.****:*. : : * *
Mflv_3325|M.gilvum_PYR-GCK EVAVVLLTGAGRAFSAGNDLKEMQAGITDGSLTADGSHFTTMIDALIDLP
Mvan_3046|M.vanbaalenii_PYR-1 EVAVVLLTGAGRAFSAGNDLKEMQAGIADGSLTSEGSHFTTMIDALTDLP
MMAR_2759|M.marinum_M QVAVVLLTGSGRGFSAGTDLAEMQARITDPNFSEGKFGFRGLIKALAGFP
MUL_2995|M.ulcerans_Agy99 QVAVVVLTGSGRGFSAGTDLAEMQARINDRNFSEGKFGFRGLIEALAGFP
MAV_2835|M.avium_104 EVAVVLLTGAGRGFSAGTDLAEMQARITDPDFTPGKHGFIGLINALSAFP
TH_3154|M.thermoresistible__bu DVSVVLLTGAGRAFSAGNDLVEMQARITDPDFTPGEHGFPGLMKALLDLR
MSMEG_3567|M.smegmatis_MC2_155 EVSVVLLTGAGRAFSSGNDLVEMQTRIANPDFTPGEHGFYGMIEALADFP
Mb1099c|M.bovis_AF2122/97 DIGAIIITGSAKAFAAGADIKEMADLTFADAFTAD---FFATWGKLAAVR
Rv1070c|M.tuberculosis_H37Rv DIGAIIITGSAKAFAAGADIKEMADLTFADAFTAD---FFATWGKLAAVR
MLBr_02402|M.leprae_Br4923 DVGAILITGSPKVFAAGADIKEMASLTFTDAFDAD---FFSAWGKLAAVR
MAB_1185c|M.abscessus_ATCC_199 GIGAIIITGSEKAFAAGADIKEMSEQSFSDMFGSD---FFSAWGKLGAVR
:..:::**: : *::* *: ** : * * .
Mflv_3325|M.gilvum_PYR-GCK KPLICAVNGVGLGIGTTILGYADLVFMSSTARLKCPFTSLGVAPEAASSY
Mvan_3046|M.vanbaalenii_PYR-1 KPLICAVNGVGLGIGTTILGYADLVFMSATARLKCPFTSLGVAPEAASSY
MMAR_2759|M.marinum_M KPLICAVNGLGVGIGATILGYADLAFMSSTARLKCPFTSLGVAPEAASSY
MUL_2995|M.ulcerans_Agy99 KPLICAVNGLGVGIGATILGYADLAFMSSTARLKCPFTSLGVAPEAASSY
MAV_2835|M.avium_104 KPLICAVNGVGVGIGATILGYADLAFMSSTARLKCPFTSLGVAPEAASSY
TH_3154|M.thermoresistible__bu KPLICAVNGVAVGIGTTILGYADLVFMADTARLKCPFTSLGVAPEAGSSY
MSMEG_3567|M.smegmatis_MC2_155 KPFLCAVNGVGVGIGATILGFADLVFMADTARLKCPFTSLGVAPEAASSY
Mb1099c|M.bovis_AF2122/97 TPTIAAVAGYALGGGCELAMMCDVLIAADTAKFGQPEIKLGVLPGMGGSQ
Rv1070c|M.tuberculosis_H37Rv TPTIAAVAGYALGGGCELAMMCDVLIAADTAKFGQPEIKLGVLPGMGGSQ
MLBr_02402|M.leprae_Br4923 TPMIAAVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGSQ
MAB_1185c|M.abscessus_ATCC_199 TPTIAAVSGYALGGGCELAMMCDVIIAAENATFGQPEIKLGVLPGMGGSQ
.* :.** * .:* * : .*: : : .* : * .*** * ..*
Mflv_3325|M.gilvum_PYR-GCK LLPQLIGRQNAAWLLLSSEWVDADEAHRMGIAWKVCEPDDLLPEARRHAE
Mvan_3046|M.vanbaalenii_PYR-1 LLPQLIGRQNAAWLLLSSEWVDAAEAERMGLAWKVCEPDDLLPEARRHAE
MMAR_2759|M.marinum_M LLPQLVGRQNAAWLLMSSEWIDAEEALRMGLVWRICSPEELLPEARRHAE
MUL_2995|M.ulcerans_Agy99 LLPQLVGRQNAAWLLMSSEWIDAEEALRMGLVWRICSPEELLPEARRHAE
MAV_2835|M.avium_104 LLPQLVGRQNAAWLLMSSEWVDADEALRMGLVWRVCAPDELLPQARHHAE
TH_3154|M.thermoresistible__bu LLPQLMGRQNAAWLLMSSEWVDAEEAQRMGIAWRVCPVDELLAEARRHAE
MSMEG_3567|M.smegmatis_MC2_155 LMPRLIGRQNAAWLLMSSEWISAAEAKEMGLVFKVCTPGDLLTEARRHAE
Mb1099c|M.bovis_AF2122/97 RLTRAIGKAKAMDLILTGRTMDAAEAERSGLVSRVVPADDLLTEARATAT
Rv1070c|M.tuberculosis_H37Rv RLTRAIGKAKAMDLILTGRTMDAAEAERSGLVSRVVPADDLLTEARATAT
MLBr_02402|M.leprae_Br4923 RLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVLADDLLPEAKAVAT
MAB_1185c|M.abscessus_ATCC_199 RLTRAIGKAKAMDMILTGRNMDAAEAERSGLVSRVVATESLLDEAKAVAK
:.: :*: :* ::::.. :.* ** . *:. :: .** :*: *
Mflv_3325|M.gilvum_PYR-GCK ILAARSIPSLIAVKKTMVAPHRAEIAAATARENAHFQELLGGVANAAALA
Mvan_3046|M.vanbaalenii_PYR-1 ILASRSIPSLIAVKKTIVEPYRAEIAAATQRENAHFQELLGGVANAAALA
MMAR_2759|M.marinum_M ILAAKPISSLMAVKHTMVEPNRAQIAAASARENAHFAELMGAQANAAALA
MUL_2995|M.ulcerans_Agy99 ILAAKPISSLMAVKHTMVEPNRAQIAAASARENAHFAELMGAQANAAALA
MAV_2835|M.avium_104 ILASRPISSLMAVKHTMMEPVRPQIAAATARENAHFAELMGAQANAAALA
TH_3154|M.thermoresistible__bu LLASRPVASLMAVKHTMVEPIRADVEAAIAREYDQFAELLGGAANADALA
MSMEG_3567|M.smegmatis_MC2_155 ILAAKPVTSLQAVKETMVAPVREAVAAATRRESELFVELVGAAANAEALA
Mb1099c|M.bovis_AF2122/97 TISQMSASAARMAKEAVNRAFESSLSEGLLYERRLFHSAFATEDQSEGMA
Rv1070c|M.tuberculosis_H37Rv TISQMSASAARMAKEAVNRAFESSLSEGLLYERRLFHSAFATEDQSEGMA
MLBr_02402|M.leprae_Br4923 TISQMSRSATRMAKEAVNRSFESTLAEGLLHERRLFHSTFVTDDQSEGMA
MAB_1185c|M.abscessus_ATCC_199 TISEMSLSASMMAKEAVNRAFESSLAEGLLFERRIFHSAFGTADQSEGMA
:: . .: .*.:: . . : . * * . . :: .:*
Mflv_3325|M.gilvum_PYR-GCK EFTQKG-------
Mvan_3046|M.vanbaalenii_PYR-1 EFTGQRKS-----
MMAR_2759|M.marinum_M DFTDRRRP-----
MUL_2995|M.ulcerans_Agy99 DFTDRRRP-----
MAV_2835|M.avium_104 DFNKR--------
TH_3154|M.thermoresistible__bu EFTGRKK------
MSMEG_3567|M.smegmatis_MC2_155 AFTGRKGAAGS--
Mb1099c|M.bovis_AF2122/97 AFIEKRAPQFTHR
Rv1070c|M.tuberculosis_H37Rv AFIEKRAPQFTHR
MLBr_02402|M.leprae_Br4923 AFIEKRAPQFTHR
MAB_1185c|M.abscessus_ATCC_199 AFVEKRPANFIHR
* :