For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DSYRRGNLEFEVSEAGPSDGATVILLHGFPQRNSSWEPIVGRLTAHGFRCLAPNQRGYSPGARPPRRRDY RVGELVEDVVALIDASGADRVHLVGHDWGAAVAWGVASYVPARLATLSALSVPHPQAFLRAMTTSRQGLA SWYMYLNQLPRLPERLMLGRDGGAARMVRTLQRSGQTREIAERDARAMAEPGALTAALNWYRAMFLSTSD PRAKHKVAKRKITVPTLYVWSDRDIAITEKPARETANYVAGPYRFETLHGVSHWIPEERPDEVADLLLEW FAAHPV*
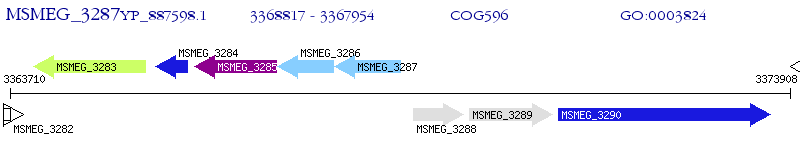
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3287 | - | - | 100% (287) | hydrolase, alpha/beta fold family protein |
| M. smegmatis MC2 155 | MSMEG_6106 | - | 2e-32 | 31.31% (313) | epoxide hydrolase |
| M. smegmatis MC2 155 | MSMEG_1998 | - | 7e-24 | 30.14% (292) | hydrolase, alpha/beta fold family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1155 | ephC | 6e-26 | 30.31% (287) | epoxide hydrolase EphC |
| M. gilvum PYR-GCK | Mflv_1415 | - | 3e-34 | 32.00% (300) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv3617 | ephA | 8e-32 | 31.21% (314) | epoxide hydrolase EphA |
| M. leprae Br4923 | MLBr_02297 | - | 4e-18 | 30.87% (298) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_0679c | - | 3e-68 | 47.72% (285) | hydrolase |
| M. marinum M | MMAR_5114 | ephA | 7e-34 | 31.72% (309) | epoxide hydrolase EphA |
| M. avium 104 | MAV_0539 | - | 9e-32 | 31.48% (305) | hydrolase, alpha/beta fold family protein |
| M. thermoresistible (build 8) | TH_0992 | ephA | 6e-29 | 27.85% (316) | PROBABLE EPOXIDE HYDROLASE EPHA (EPOXIDE HYDRATASE) |
| M. ulcerans Agy99 | MUL_4193 | ephA | 2e-34 | 31.82% (308) | epoxide hydrolase EphA |
| M. vanbaalenii PYR-1 | Mvan_5373 | - | 6e-36 | 33.55% (310) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3287|M.smegmatis_MC2_155 ------------------------MDSYRRGNLEFEVSEAG---------
MAB_0679c|M.abscessus_ATCC_199 ---------------------MSRLSTFTNDGLTFDVIDEG---------
MMAR_5114|M.marinum_M ------------------MVATSTERLVDTNGVRLRVTEAG---------
MUL_4193|M.ulcerans_Agy99 ------------------MVATSTERLVDTNGVRLRVTEAG---------
Rv3617|M.tuberculosis_H37Rv -------------------MGAPTERLVDTNGVRLRVVEAG---------
MAV_0539|M.avium_104 --------------------------------MRLRVTEAG---------
TH_0992|M.thermoresistible__bu -------------------VITPSERSVETNGVRLRLVEAG---------
Mflv_1415|M.gilvum_PYR-GCK ----------------------MQSRMVHTNGITLRVFEAG---------
Mvan_5373|M.vanbaalenii_PYR-1 MNSRWCFRAVRDRGCPFGESGAMRSRMVQTNGVTLRVTEAG---------
MLBr_02297|M.leprae_Br4923 --------MAMSNPSVTRIAGPWHHLDVHANGIRFHVVEAVPTQPAGQDC
Mb1155|M.bovis_AF2122/97 ----MRAGRGERESTWRTTMAEPHWIDVKGPNGDLKALTWG---------
:
MSMEG_3287|M.smegmatis_MC2_155 ---PSDGATVILLHGFPQRNSSWEPIVGRLTAHGFRCLAPNQRGYSPGAR
MAB_0679c|M.abscessus_ATCC_199 ---ALDGPVIVALHGFPERATMWEEVIPQLTGAGFRVLAPNQRGYSPGAR
MMAR_5114|M.marinum_M ---ERGAPVVILAHGFPELAYSWRHQIPVLAEAGYHVLAPDQRGYGGSSR
MUL_4193|M.ulcerans_Agy99 ---ERGAPVVILAHGFPELAYSWRHQIPVLAEAGYHVLAPDQRGYGGSSR
Rv3617|M.tuberculosis_H37Rv ---EPGAPVVILAHGFPELAYSWRHQIPALADAGYHVLAPDQRGYGGSSR
MAV_0539|M.avium_104 ---DRGAPVVILAHGFPELAYSWRHQIDVLADAGYHVLAPDQRGYGGSDR
TH_0992|M.thermoresistible__bu ---ERGDPLVVLAHGFPELAYSWRHQIPALVDAGYHVMAPDQRGYGGSSA
Mflv_1415|M.gilvum_PYR-GCK ---ERSAPVVVLCHGFPELAFTWRHQISALAAAGFHVLAPDQRGYGGSDK
Mvan_5373|M.vanbaalenii_PYR-1 ---EPGAPVVVLCHGFPELAFTWRHQVRALADAGFHVLAPDQRGYGGSDK
MLBr_02297|M.leprae_Br4923 APSATARPLVMLLHGFGSFWWSWRHQLRGLTEA--RLVAVDLRGYGGSDK
Mb1155|M.bovis_AF2122/97 ---PAGAPVALCLHGFPDTAYGWRKVAPRLAESGWHVVAPFMRGYAPSSI
. : *** . *. *. : :* ***. .
MSMEG_3287|M.smegmatis_MC2_155 PPRRRDYRVGELVEDVVALIDASG-ADRVHLVGHDWGAAVAWGVASYVPA
MAB_0679c|M.abscessus_ATCC_199 PVGVRNYAVDKLEGDIIALVDQAG-VRKFHVLAHDWGAAVAWQLAGNHAD
MMAR_5114|M.marinum_M PEAIEDYDIHQLTADLVGLLDDVG-AERAAWVGHDWGAVVVWNAPLLHPD
MUL_4193|M.ulcerans_Agy99 PEAIEDYDIHQLTADLVGLLDDVG-AERAAWVGHDWGAVVVWNAPLLHPD
Rv3617|M.tuberculosis_H37Rv PEAIEAYDIHRLTADLVGLLDDVG-AERAVWVGHDWGAVVVWNAPLLHAD
MAV_0539|M.avium_104 PDAVEAYDIHQLTADLVGLLDDVG-AQRAVWVGHDWGAAVVWNAPLLHPD
TH_0992|M.thermoresistible__bu PEAIEAYDITRLTADLMGLLDDIG-AEKAAFIGHDWGALVVWNAALLYPD
Mflv_1415|M.gilvum_PYR-GCK PGDAGVYNVAELTADVVGLLDDVG-AERAALVGHDFGAVVAWGAPLLEPD
Mvan_5373|M.vanbaalenii_PYR-1 PDAVDSYNVAELTADVVGLLDDLG-AERAALVGHDFGAVVAWAAPLLQPD
MLBr_02297|M.leprae_Br4923 PP--RGYDGWTLAGDTAGLIRALG-HSSATLVGHAEGGLACWATALLHPR
Mb1155|M.bovis_AF2122/97 PAD-GSYHVGALMHDALRVRSAAGGTERDVIIGHDWGAIAATGLAAMPDS
* * * * : * :.* *. . .
MSMEG_3287|M.smegmatis_MC2_155 RLATLSALSVPHPQAFL------RAMTTSRQGLAS----WYMYLNQLPRL
MAB_0679c|M.abscessus_ATCC_199 RVQTLSILSVPHPRAFQ------DAMLRG-QLLHS----WYMLLFQIPRL
MMAR_5114|M.marinum_M RVAAVAGLSVPVLPRAQ------VPPIQAFRQRFG-EHFFYILYFQQPGV
MUL_4193|M.ulcerans_Agy99 RVAAVAGLSVPVLPRAQ------VPPIQAFRQRFG-EHFFYILYFQQPGV
Rv3617|M.tuberculosis_H37Rv RVAAVAALSVPALPRAQ------VPPTQAFRSRFG-ENFFYILYFQEPGI
MAV_0539|M.avium_104 RVAAVAAMSVPVTPRPR------LAPTQAWRKMFG-ENFFYILYFQEPGV
TH_0992|M.thermoresistible__bu RVAAVAGLSVPPVPRSL------TRPTEAFRALVGEDNFFYILYFQEPGV
Mflv_1415|M.gilvum_PYR-GCK RFSAVAGLSLPPVPRPQ------VPTTQAFRRVFG-DRFMYILYFQEPGP
Mvan_5373|M.vanbaalenii_PYR-1 RFSSVAGLSLPPVPRPK------VPTTQAFRRIFA-DRFFYILYFQDRGP
MLBr_02297|M.leprae_Br4923 LVRAIALINSPHPAALR------RSMLTHRNQGYA--LLPTLLRYQLPIW
Mb1155|M.bovis_AF2122/97 PFAKAVIMSVPPSAAFRPLGRVPERGRLLRELPHQLLRSWYILYFQLPWL
. :. * . : *
MSMEG_3287|M.smegmatis_MC2_155 PERLMLGRDGGAARMVRTLQRSGQTREIAERDA--------RAMAEPG--
MAB_0679c|M.abscessus_ATCC_199 PEWLFGFNETEAFKKFGKS-WMKVSDRLAERIR--------ELFSEPG--
MMAR_5114|M.marinum_M ADAELNGDPARTMRRMIGGQRSPTDQGAALRMVAPGPEGFIDRLPEPDTP
MUL_4193|M.ulcerans_Agy99 ADAELNGDPARTMRRMIGGQRSPTDQGAALRMVAPGPEGFIDRLPEPDAP
Rv3617|M.tuberculosis_H37Rv ADAELNGDPARTMRRMIGGLRPPGDQSAAMRMLAPGPDGFIDRLPEPAGL
MAV_0539|M.avium_104 ADAELNADPAQVMRRMMGSLRTDGDKAAGLRMVAPGPEGFLERLPEPDGL
TH_0992|M.thermoresistible__bu ADAELDGDPARTMRRMFGGLTSDPD--AAHRMLQPGPAGFIDRLPEPEAL
Mflv_1415|M.gilvum_PYR-GCK ADAELARDPATTFRRLFA--LTTG---GAEMVGDAGPQGFLDRIPEPGGL
Mvan_5373|M.vanbaalenii_PYR-1 ADAELARDPAGTFRRLFA--MDVADEQSALRMTEPGPAGFLDRIPDPGRL
MLBr_02297|M.leprae_Br4923 PERLLTRNNAAEIERLVRSRSSAKWQACEDFAVTIDHLRIAIQIPAAAHC
Mb1155|M.bovis_AF2122/97 PERSASWVVPLLWRRWSPGYHAEEDLRHVDAAIGTPEGRRAALGPYRATM
.: . .
MSMEG_3287|M.smegmatis_MC2_155 -------------------ALTAALNWYRAMFLSTSDPRAKHKVAKRKIT
MAB_0679c|M.abscessus_ATCC_199 -------------------ATTATVNWYRAMRYSLRSP-------ARPSR
MMAR_5114|M.marinum_M PSWISRDELDHYISEFTRTGFTGGLNWYRNFDRNWETT---ADLEGATIT
MUL_4193|M.ulcerans_Agy99 PSWISRDELDHYISEFTRTGFTGGLNWYRNFDRNWETT---ADLEGATIT
Rv3617|M.tuberculosis_H37Rv PAWISQEELDHYIGEFTRTGFTGGLNWYRNFDRNWETT---ADLAGKTIS
MAV_0539|M.avium_104 PEWISQDELDHYIAEFSRTGFTGGLNWYRNFDRNWETT---PELDGAKIA
TH_0992|M.thermoresistible__bu PDWLTAEELDHYIAEFTRTGFTGGLNWYRNMDRNWELT---EHLAGATIT
Mflv_1415|M.gilvum_PYR-GCK PDWISQADFDVYVDEFTRGGFTGPLNWYRCFDRNWELT---AETPAPTIE
Mvan_5373|M.vanbaalenii_PYR-1 PDWISPPDFAVYVDEFRRGGFTAPLNWYRCFDLNWELT---ADPPAPTIG
MLBr_02297|M.leprae_Br4923 ALEYQR---------------WAVRSQLRNEGRKFMKS------MSRPFS
Mb1155|M.bovis_AF2122/97 RN-------------------TRAPADYADLNRLWTEAP----------K
.
MSMEG_3287|M.smegmatis_MC2_155 VPTLYVWSDRDIAITEKPARETANYVAGPYRFETLHGVSHWIPEERPDEV
MAB_0679c|M.abscessus_ATCC_199 VPTLFIWSDGDSAVTRAAADATPKYVEAPYRFEVFEGVSHWIAEERPLET
MMAR_5114|M.marinum_M VPSLFMAGTKDPVLSFARSDRASEVITGPYREVMVEGAGHWLQQERPDEV
MUL_4193|M.ulcerans_Agy99 VPSLFMAGTKDPVLSFARSDRASEVITGPYREVMVEGAGHWLQQERPDEV
Rv3617|M.tuberculosis_H37Rv VPSLFIAGTADPVLTFTRTDRAAEVISGPYREVLIDGAGHWLQQERPGEV
MAV_0539|M.avium_104 VPCLFIGGTADPVLSFTRADRAAEVISGPYRHVMIDGAGHWLQQERPAEV
TH_0992|M.thermoresistible__bu APALFLAGAADPVLGFMRPERATEVAVGPYRQVLLDGAGHWVQQERPQEV
Mflv_1415|M.gilvum_PYR-GCK VPALFVGGTEDATLAYTPRDRVREVVTGDYREVMIDGAGHWLTEERPDEV
Mvan_5373|M.vanbaalenii_PYR-1 VPALFVGGTADATLAYTPRHRVREVVTGDYREVMIDGAGHWLTEERPDEV
MLBr_02297|M.leprae_Br4923 VPLLHVRGDADPYLLADSVDRTRRYAP-HGRYVSVSDAGHFSHEEAPEEI
Mb1155|M.bovis_AF2122/97 LPVLYLHGHDDGCATSAFTHWTARVLPAGSEVAVVEHAGHFLQLEQPDKI
* *.: . * . . . . ..*: * * :
MSMEG_3287|M.smegmatis_MC2_155 ADLLLEWFAAHPV----
MAB_0679c|M.abscessus_ATCC_199 AALVLEHIAEYSA----
MMAR_5114|M.marinum_M NATLLDFLKGVQW----
MUL_4193|M.ulcerans_Agy99 NATLLDFLKGVQW----
Rv3617|M.tuberculosis_H37Rv TAALLEFLTGLELR---
MAV_0539|M.avium_104 NAALLDFLNGLELR---
TH_0992|M.thermoresistible__bu NAALIDFLRGLELQ---
Mflv_1415|M.gilvum_PYR-GCK SRVLVDFLTTVR-----
Mvan_5373|M.vanbaalenii_PYR-1 SRILLEFLTAAR-----
MLBr_02297|M.leprae_Br4923 NRYLTRFLKQVHGPRLS
Mb1155|M.bovis_AF2122/97 AELIVAFIGSPG-----
: :