For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MFNRTELLPGQKRWSIGSVAADVGLVFWFVFSLFPIFWMLMLALKNAEEQTTTYFAFSPTWENFATVLTD KGTEMTSVDFKASLLTSLLNCGGAVIVSLLIGIPAAYAAGRWQYKGSNDLMFTMLSFRFAPELMVIVPLF VIYNQIGLFDTKVGMIWVLQLVTMPLVVWILRSYFQDLPEDLEQAALLDGYTRRRAFLMVALPIVRPGIA AAALLAFIFAWNNYVFPLILADSNAGTVTVAITKFLGGGGQAYYNLTAAAAIIAALPPLILALTIQRYLV RGLSFGAVKA
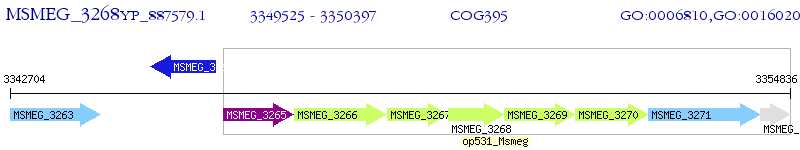
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3268 | - | - | 100% (290) | ABC transporter, permease protein |
| M. smegmatis MC2 155 | MSMEG_5572 | - | 3e-43 | 37.45% (275) | sugar ABC transporter permease protein |
| M. smegmatis MC2 155 | MSMEG_3109 | - | 4e-36 | 35.88% (262) | binding-protein-dependent transport systems inner membrane |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1269 | sugB | 2e-34 | 32.96% (267) | sugar-transport integral membrane protein ABC transporter |
| M. gilvum PYR-GCK | Mflv_3853 | - | 4e-36 | 30.39% (283) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv1237 | sugB | 2e-34 | 32.96% (267) | sugar-transport integral membrane protein ABC transporter |
| M. leprae Br4923 | MLBr_01088 | - | 2e-35 | 33.84% (263) | putative ABC-transport protein, inner membrane component |
| M. abscessus ATCC 19977 | MAB_1374 | - | 2e-34 | 32.44% (262) | sugar ABC transporter, permease protein SugB |
| M. marinum M | MMAR_4204 | sugB | 2e-35 | 34.22% (263) | sugar-transport integral membrane protein ABC transporter |
| M. avium 104 | MAV_1376 | sugB | 6e-34 | 33.33% (264) | ABC transporter, permease protein SugB |
| M. thermoresistible (build 8) | TH_2009 | - | 5e-42 | 36.67% (270) | sugar ABC transporter permease protein |
| M. ulcerans Agy99 | MUL_4511 | sugB | 1e-29 | 34.43% (212) | sugar-transport integral membrane protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_2542 | - | 8e-36 | 30.39% (283) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4204|M.marinum_M -------------------MTTGSGRRTFWVVIDTMVVVYALLPVLWILS
MUL_4511|M.ulcerans_Agy99 ------------------------------------------------MS
Mb1269|M.bovis_AF2122/97 ---------------------MGARRATYWAVLDTLVVGYALLPVLWIFS
Rv1237|M.tuberculosis_H37Rv ---------------------MGARRATYWAVLDTLVVGYALLPVLWIFS
MLBr_01088|M.leprae_Br4923 --------MGIEKVKVTAGVRVGARRATIWVIIDTFVVGYALLPVLWILS
MAV_1376|M.avium_104 -------------------------------MIDTLVVVYALLPVLWIFS
MAB_1374|M.abscessus_ATCC_1997 ------------------MSRPDSSRTAGWLIADVLVLCYALVPVLWVLS
Mflv_3853|M.gilvum_PYR-GCK ----------------------MTKNTKWWTIAGIVIVIYSFVPMLWMIS
Mvan_2542|M.vanbaalenii_PYR-1 ----------------------MTKNTKWWTIAGIVIVIYSFVPMLWMIS
MSMEG_3268|M.smegmatis_MC2_155 ----MFNRTELLPGQ--KRWSIGSVAAD-VGLVFWFVFS--LFPIFWMLM
TH_2009|M.thermoresistible__bu MSTATVTTTAPVPTEPSKAAKVKRRSFDPWGVVAWIAAIGFFFPVFWMVL
.
MMAR_4204|M.marinum_M LSLKPTSTVKDGR--LIPSSVTLDNYRGVFGGD-------LFSSALINSI
MUL_4511|M.ulcerans_Agy99 LSPKPTSTVKDGR--LIPSSVTLDNYRGVFGGD-------LFSSALINSI
Mb1269|M.bovis_AF2122/97 LSLKPTSTVKDGK--LIPSTVTFDNYRGIFRGD-------LFSSALINSI
Rv1237|M.tuberculosis_H37Rv LSLKPTSTVKDGK--LIPSTVTFDNYRGIFRGD-------LFSSALINSI
MLBr_01088|M.leprae_Br4923 LSLKPASTVKDGK--LIPSLVTFDNYRGIFRSD-------LFSSALINSI
MAV_1376|M.avium_104 LSLKPTSTVKDGK--LIPSAISLENYRGIFRGD-------FFSSALINSV
MAB_1374|M.abscessus_ATCC_1997 LSLKPTSSVKDGK--FFPWPITLDNYRGIFSGN-------VFTSALVNSI
Mflv_3853|M.gilvum_PYR-GCK LAFKPPSDIVSGEPSFLPSTVTFDNFAQIFSNP-------LFTRALINSI
Mvan_2542|M.vanbaalenii_PYR-1 LAFKPPSDIVSGKPSFLPSSITFDNFAQIFSQP-------LFTRALINSI
MSMEG_3268|M.smegmatis_MC2_155 LALKNAEEQTTTY---FAFSPTWENFATVLTDKGTEMTSVDFKASLLTSL
TH_2009|M.thermoresistible__bu TAFKQEVDAYTTTP-KVFFAPTLDQFRAVFDQG--------IGTALLNSL
: * . : ::: :: : :*:.*:
MMAR_4204|M.marinum_M GIGLTTTVIAVLLGAMAAYAVARLDFPGKRLLVGVALLITMFPAISLVTP
MUL_4511|M.ulcerans_Agy99 GIGLTTTVIAVLLGAMAACAVARLDFPGKRLLVGVTLLITMFPAISLVTP
Mb1269|M.bovis_AF2122/97 GIGLITTVIAVVLGAMAAYAVARLEFPGKRLLIGAALLITMFPSISLVTP
Rv1237|M.tuberculosis_H37Rv GIGLITTVIAVVLGAMAAYAVARLEFPGKRLLIGAALLITMFPSISLVTP
MLBr_01088|M.leprae_Br4923 GIGLTTTVIAVMFGAMAAYAIARLAFPGKRLLVGTTLLVTMFPAISLVTP
MAV_1376|M.avium_104 GIGLITTAVAVLLGAMAAYAVARLDFPGKRLLIGATLLITMFPAISLVTP
MAB_1374|M.abscessus_ATCC_1997 GIGLIATVIAVSVGTMAAYAVARLDFPGKKALIGAALLIAMFPQISLVTP
Mflv_3853|M.gilvum_PYR-GCK GIALVATVISVIIAMFAAYAIARLEFPGKKVLLSLALGIAMFPQAALVGP
Mvan_2542|M.vanbaalenii_PYR-1 GIAVIATVISVIIAMFAAYAIARLEFPGKKVLLSLALGIAMFPQAALVGP
MSMEG_3268|M.smegmatis_MC2_155 LNCGGAVIVSLLIGIPAAYAAGRWQYKGSNDLMFTMLSFRFAPELMVIVP
TH_2009|M.thermoresistible__bu FATVVSTLLVLALGVPAAFALSLRPVRKTQDALFFFMSTKMLPIVAAIIP
:. : : .. ** * . .. : : : * : *
MMAR_4204|M.marinum_M LFNIERRIGLFDTWPGLILPYITFALPLAIYTLSAFFREIPWDLEKAAQM
MUL_4511|M.ulcerans_Agy99 LFNIERRIGLFDTWPELILPYITFALPLAIYTLSAFFREIPWDLEKAAQM
Mb1269|M.bovis_AF2122/97 LFNIERAIGLFDTWPGLILPYITFALPLAIYTLSAFFREIPWDLEKAAKM
Rv1237|M.tuberculosis_H37Rv LFNIERAIGLFDTWPGLILPYITFALPLAIYTLSAFFREIPWDLEKAAKM
MLBr_01088|M.leprae_Br4923 LFNIERRVGLFDTWAGLILPYITFALPLAIYTLSAFFAEIPWDLEKAAKM
MAV_1376|M.avium_104 LFNIERFLGLFDTWPGLILPYITFALPLAIYTLSAFFREIPWDLEKAAKI
MAB_1374|M.abscessus_ATCC_1997 IFNIERSVGLFDTWPGLIIPYITFALPLAIYTLSAFFREIPWELEKAAKM
Mflv_3853|M.gilvum_PYR-GCK LFDMWRGLGIYDTWLGLIIPYLTFALPLSIWTMSAFFRQIPWEMEHAAQV
Mvan_2542|M.vanbaalenii_PYR-1 LFDMWRGLGIYDTWLGLIIPYLTFALPLSIWTMSAFFRQIPWEMEHAAQV
MSMEG_3268|M.smegmatis_MC2_155 LFVIYNQIGLFDTKVGMIWVLQLVTMPLVVWILRSYFQDLPEDLEQAALL
TH_2009|M.thermoresistible__bu LYVIVSNVGLLDNIWALIILYTAMNLPIAVWMMRSFFLEVPGELLEAASL
:: : :*: *. :* . :*: :: : ::* ::* :: .** :
MMAR_4204|M.marinum_M DGATPYQAFRKVIVPLAAPGLVTAAILVFIFAWNDLLLALSLTATKASIT
MUL_4511|M.ulcerans_Agy99 DGATPYQAFRKVIVPLAAPGLVTAAILVFIFAWNDLLLALSLTATKASIT
Mb1269|M.bovis_AF2122/97 DGATPGQAFRKVIVPLAAPGLVTAAILVFIFAWNDLLLALSLTATKAAIT
Rv1237|M.tuberculosis_H37Rv DGATPGQAFRKVIVPLAAPGLVTAAILVFIFAWNDLLLALSLTATKAAIT
MLBr_01088|M.leprae_Br4923 DGATSGQAFRKVIVPLAAPGLVTAAILVFIFAWNDLLLALSLTSTKAAIT
MAV_1376|M.avium_104 DGATPAQAFRKVIVPLAAPGVVTAAILVFIFAWNDLLLALTLTATKAAIT
MAB_1374|M.abscessus_ATCC_1997 DGATPAQAFWKVIAPLATPGIVTSAILVFIFAWNDLLLAISLTATDRSIT
Mflv_3853|M.gilvum_PYR-GCK DGATQWQAFRKVIVPLAAPGVFTTAILTFFFCWNDFLFAISLTSTDSART
Mvan_2542|M.vanbaalenii_PYR-1 DGATQWQAFRKVIVPLAAPGVFTTAILTFFFCWNDFLFAISLTSTDSART
MSMEG_3268|M.smegmatis_MC2_155 DGYTRRRAFLMVALPIVRPGIAAAALLAFIFAWNNYVFPLILADSNAGTV
TH_2009|M.thermoresistible__bu DGAGTWRTMREVILPIVSPGIAATALICVIFAWNEFFFAVNLTAVQAQTM
** ::: * *:. **: ::*:: .:*.**: .:.: *: .
MMAR_4204|M.marinum_M APVAIANFAGSSQFEEPTGSIAAGAMVITVPIIVFVLIFQRRIVAGLTSG
MUL_4511|M.ulcerans_Agy99 APIAIANFAGSSQFEEPTGSIAAGAMVITVPIIVFVLIFQLRIVAGLTSG
Mb1269|M.bovis_AF2122/97 APVAIANFTGSSQFEEPTGSIAAGAIVITIPIIVFVLIFQRRIVAGLTSG
Rv1237|M.tuberculosis_H37Rv APVAIANFTGSSQFEEPTGSIAAGAIVITIPIIVFVLIFQRRIVAGLTSG
MLBr_01088|M.leprae_Br4923 APVAITNFTGSSQFEEPTGSIAAGAIVITAPIIVFVWIFQRRIVAGLTSG
MAV_1376|M.avium_104 APVAIVSFSGSSQFEEPTGSIAAGAVVITVPVILFVLIFQRRIVAGLTSG
MAB_1374|M.abscessus_ATCC_1997 APVAIANFTGSSQFEEPTGSIAAGAIVITVPIIIFVLIFQRRIVAGLTSG
Mflv_3853|M.gilvum_PYR-GCK VPAALAFFQGASYFESPVPYIMAASVIVTIPVVILVLVFQRRIVAGLTSG
Mvan_2542|M.vanbaalenii_PYR-1 VPAALAFFQGASYFESPVPYIMAASVIVTIPVVILVLIFQRRIVAGLTSG
MSMEG_3268|M.smegmatis_MC2_155 TVAITKFLGGGGQAYYNL--TAAAAIIAALPPLILALTIQRYLVRGLSFG
TH_2009|M.thermoresistible__bu PVFLVGFIAGEG-LYWAK--LCAASTMAALPVILAGWLAQNKLVRGLSFG
: * . *.: : : * :: * :* **: *
MMAR_4204|M.marinum_M AVKG
MUL_4511|M.ulcerans_Agy99 AVKG
Mb1269|M.bovis_AF2122/97 AVKG
Rv1237|M.tuberculosis_H37Rv AVKG
MLBr_01088|M.leprae_Br4923 AVKG
MAV_1376|M.avium_104 AVKG
MAB_1374|M.abscessus_ATCC_1997 AVKG
Mflv_3853|M.gilvum_PYR-GCK AVKG
Mvan_2542|M.vanbaalenii_PYR-1 AVKG
MSMEG_3268|M.smegmatis_MC2_155 AVKA
TH_2009|M.thermoresistible__bu AIK-
*:*