For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNNGGLQAEVHTLERAIFEVKRIIVGQDQLVERMLVGLLAKGHVLLEGVPGVAKTLAVETFAKVVGGTFA RIQFTPDLVPTDIVGTRIYRQGKEEFEIELGPAVVNFLLADEINRAPAKVQSALLEIMAERKISIGGQTF PLPSPFLVMATQNPIEQEGVYQLPEAQRDRFLFKLNVDYPSPEEEREIIYRMGVKPPEPKQILTTGDLLR LQDVAANTFVHHALVDYVVRIVTATREPEKFGMPDAKAWIAYGASPRASLGIISAARALALVRGRDYVIP QDVVEVIPDVLRHRLVLTYDALADEISSETVVNRILQTVALPQVNALPQQGHSVPPAVPAAAAAASGR
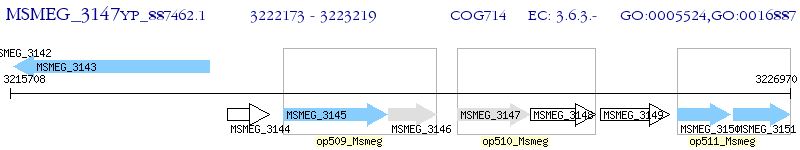
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3147 | moxR | - | 100% (348) | ATPase, MoxR family protein |
| M. smegmatis MC2 155 | MSMEG_6612 | moxR | 1e-75 | 46.86% (303) | ATPase, MoxR family protein |
| M. smegmatis MC2 155 | MSMEG_6219 | - | 9e-66 | 43.87% (310) | ATPase family protein associated with various cellular |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1515 | moxR1 | 1e-169 | 86.05% (344) | transcriptional regulatory protein MOXR1 |
| M. gilvum PYR-GCK | Mflv_3661 | - | 0.0 | 96.53% (346) | ATPase |
| M. tuberculosis H37Rv | Rv1479 | moxR1 | 1e-169 | 86.05% (344) | transcriptional regulatory protein MOXR1 |
| M. leprae Br4923 | MLBr_01810 | moxR | 1e-168 | 85.47% (344) | MoxR homolog |
| M. abscessus ATCC 19977 | MAB_2726c | - | 1e-160 | 81.56% (347) | hypothetical protein MAB_2726c |
| M. marinum M | MMAR_2286 | moxR1 | 1e-171 | 87.06% (340) | transcriptional regulatory protein, MoxR1 |
| M. avium 104 | MAV_3299 | - | 1e-170 | 86.92% (344) | MoxR protein |
| M. thermoresistible (build 8) | TH_3229 | - | 1e-176 | 90.41% (344) | PUTATIVE ATPase associated with various cellular activities, |
| M. ulcerans Agy99 | MUL_1488 | moxR1 | 1e-171 | 87.06% (340) | transcriptional regulatory protein, MoxR1 |
| M. vanbaalenii PYR-1 | Mvan_2749 | - | 0.0 | 95.39% (347) | ATPase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3661|M.gilvum_PYR-GCK -----MTSPSGPPQGAGGYPGQGPAQGYSAGAHAAGNSAPGSAQTNGGLQ
Mvan_2749|M.vanbaalenii_PYR-1 -----MTSPSGPPQGAGGYPGQVPAQGYSSGAHAAP-SQPGPPQSNGGLQ
MSMEG_3147|M.smegmatis_MC2_155 -------------------------------------------MNNGGLQ
TH_3229|M.thermoresistible__bu VEELRMTSPSGPQQGSGGYSGSAPAPGYPAGSHAAPPTNGG----QSGLQ
MMAR_2286|M.marinum_M -----MTSAGG------GYSGPGGHS--APPAHEAPSGGP------DGLA
MUL_1488|M.ulcerans_Agy99 -----MTSAGG------GYSGPGGHS--APPAHEAPSGGP------DGLA
Mb1515|M.bovis_AF2122/97 -----MTSAGGFPAGAGGYQTPGGHS--ASPAHEAPPGGA------EGLA
Rv1479|M.tuberculosis_H37Rv -----MTSAGGFPAGAGGYQTPGGHS--ASPAHEAPPGGA------EGLA
MLBr_01810|M.leprae_Br4923 -----MTSAGGLPVGADGCSGPGGQS--GPRAHEAPPGGA------DGLV
MAV_3299|M.avium_104 -----MTAAGGPPPGAGGYSGPGGQS--GPGAHEAPTGGAG---AGNGLA
MAB_2726c|M.abscessus_ATCC_199 ------MSSPGQPGGGAGYTGGSAGS-----APHAPQGATS-----PALA
.*
Mflv_3661|M.gilvum_PYR-GCK NEVHTLERAIFEVKRIIVGQDQLVERMLVGLLAKGHVLLEGVPGVAKTLA
Mvan_2749|M.vanbaalenii_PYR-1 NEVHTLERAIFEVKRIIVGQDQLVERMLVGLLAKGHVLLEGVPGVAKTLA
MSMEG_3147|M.smegmatis_MC2_155 AEVHTLERAIFEVKRIIVGQDQLVERMLVGLLAKGHVLLEGVPGVAKTLA
TH_3229|M.thermoresistible__bu ADVHTLERTIFEVKRIIVGQDQLVERMLVGLLAKGHVLLEGVPGVAKTLA
MMAR_2286|M.marinum_M AEVHTLERAIFEVKRIIVGQDQLVERMLVGLLAKGHVLLEGVPGVAKTLA
MUL_1488|M.ulcerans_Agy99 AEVHTLERAIFEVKRIIVGQDQLVERMLVGLLAKGHVLLEGVPGVAKTLA
Mb1515|M.bovis_AF2122/97 AEVHTLERAIFEVKRIIVGQDQLVERMLVGLLSKGHVLLEGVPGVAKTLA
Rv1479|M.tuberculosis_H37Rv AEVHTLERAIFEVKRIIVGQDQLVERMLVGLLSKGHVLLEGVPGVAKTLA
MLBr_01810|M.leprae_Br4923 AEVHTLERAIFEVKRVIVGQDQLVERMLVGLLSKGHVLLEGVPGVAKTLA
MAV_3299|M.avium_104 AEVQTLERAIFEVKRIIVGQDQLVERMLVGLLSKGHVLLEGVPGVAKTLA
MAB_2726c|M.abscessus_ATCC_199 AEVQNLERAIFEVKRIIVGQDRLVERILVGLLAKGHVLLEGVPGVAKTLA
:*:.***:******:*****:****:*****:*****************
Mflv_3661|M.gilvum_PYR-GCK VETFAKVVGGTFARIQFTPDLVPTDIVGTRIYRQGKEEFDIELGPVVVNF
Mvan_2749|M.vanbaalenii_PYR-1 VETFAKVVGGTFARIQFTPDLVPTDIIGTRIYRQGKEEFDIELGPVVVNF
MSMEG_3147|M.smegmatis_MC2_155 VETFAKVVGGTFARIQFTPDLVPTDIVGTRIYRQGKEEFEIELGPAVVNF
TH_3229|M.thermoresistible__bu VETFARVVGGTFARIQFTPDLVPTDIVGTRIYRQGKEEFDIELGPVMVNF
MMAR_2286|M.marinum_M VETFARVVGGSFARIQFTPDLVPTDIVGTRIYRQGKEEFDTELGPVVANF
MUL_1488|M.ulcerans_Agy99 VETFARVVGGSFARIQFTPDLVPTDIVGTRIYRQGKEEFDTELGPVVANF
Mb1515|M.bovis_AF2122/97 VETFARVVGGTFSRIQFTPDLVPTDIIGTRIYRQGREEFDTELGPVVANF
Rv1479|M.tuberculosis_H37Rv VETFARVVGGTFSRIQFTPDLVPTDIIGTRIYRQGREEFDTELGPVVANF
MLBr_01810|M.leprae_Br4923 VETFARVVGGTFARIQFTPDLVPTDIIGTRIYRQGKEEFDTELGPVVVNF
MAV_3299|M.avium_104 VETFAKVVGGTFARIQFTPDLVPTDIIGTRIYRQGKEEFDTELGPVVVNF
MAB_2726c|M.abscessus_ATCC_199 VETFAKVVGGTFSRIQFTPDLVPTDIIGTRIYRQGREEFDTELGPVVANF
*****:****:*:*************:********:***: ****.:.**
Mflv_3661|M.gilvum_PYR-GCK LLADEINRAPAKVQSALLEIMAERKISIGGKTFPLPSPFLVMATQNPIEQ
Mvan_2749|M.vanbaalenii_PYR-1 LLADEINRAPAKVQSALLEVMAERKISIGGKTFPLPSPFLVMATQNPIEQ
MSMEG_3147|M.smegmatis_MC2_155 LLADEINRAPAKVQSALLEIMAERKISIGGQTFPLPSPFLVMATQNPIEQ
TH_3229|M.thermoresistible__bu LLADEINRAPAKVQSALLEVMAERKISIGGKTFLLPKPFLVMATQNPIEQ
MMAR_2286|M.marinum_M LLADEINRAPAKVQSALLEVMAERHVSIGGKTFPMPNPFLVMATQNPIEQ
MUL_1488|M.ulcerans_Agy99 LLADEINRAPAKVQSALLEVMAERHVSIGGKTFPMPNPFLVMATQNPIEQ
Mb1515|M.bovis_AF2122/97 LLADEINRAPAKVQSALLEVMQERHVSIGGRTFPMPSPFLVMATQNPIEH
Rv1479|M.tuberculosis_H37Rv LLADEINRAPAKVQSALLEVMQERHVSIGGRTFPMPSPFLVMATQNPIEH
MLBr_01810|M.leprae_Br4923 LLADEINRAPAKVQSALLEVMQERHVSIGGKTFPLPNPFLVMATQNPIEH
MAV_3299|M.avium_104 LLADEINRAPAKVQSALLEVMQERHVSIGGKTFPLPNPFLVMATQNPIEH
MAB_2726c|M.abscessus_ATCC_199 LLADEINRAPAKVQSALLEVMAERKISIGGKDYHLPKPFLVMATQNPIEN
*******************:* **::****: : :*.************:
Mflv_3661|M.gilvum_PYR-GCK EGVYQLPEAQRDRFLFKLNVDYPSPEEEREIIYRMGVKPPEPKGILTTAD
Mvan_2749|M.vanbaalenii_PYR-1 EGVYQLPEAQRDRFLFKLNVDYPTPEEEREIIYRMGVKPPEPKAILNTGD
MSMEG_3147|M.smegmatis_MC2_155 EGVYQLPEAQRDRFLFKLNVDYPSPEEEREIIYRMGVKPPEPKQILTTGD
TH_3229|M.thermoresistible__bu EGVYALPEAQRDRFLFKLNIDYPSPEEEREIIYRMGVKPPEPKQVLAPGD
MMAR_2286|M.marinum_M EGVYPLPEAQRDRFLFKINVGYPTPEEEREIIYRMGVTPPQPKQILGTGD
MUL_1488|M.ulcerans_Agy99 EGVYPLPEAQRDRFLFKINVGYPTPEEEREIIYRMGVTPPQPKQILGTGD
Mb1515|M.bovis_AF2122/97 EGVYPLPEAQRDRFLFKINVGYPSPEEEREIIYRMGVTPPQAKQILSTGD
Rv1479|M.tuberculosis_H37Rv EGVYPLPEAQRDRFLFKINVGYPSPEEEREIIYRMGVTPPQAKQILSTGD
MLBr_01810|M.leprae_Br4923 EGVYLLPEAQRDRFLFKINVGYPSPEEEREIIYRMGVTPPVPKQILNAGD
MAV_3299|M.avium_104 EGVYPLPEAQRDRFLFKINVGYPSPEEEREIIYRMGVRPPEPKQILNTGD
MAB_2726c|M.abscessus_ATCC_199 EGVYPLPEAQRDRFLFKINVDYPTVEEEREIIYRMGVTPPEPKQILSSEA
**** ************:*:.**: ************ ** .* :* .
Mflv_3661|M.gilvum_PYR-GCK LLRLQDVAANNFVHHALVDYVVRIVTATREPEKFGMPDAKAWIAYGASPR
Mvan_2749|M.vanbaalenii_PYR-1 LLRLQDVAANNFVHHALVDYVVRIVTATREPEKFGMPDAKAWIAYGASPR
MSMEG_3147|M.smegmatis_MC2_155 LLRLQDVAANTFVHHALVDYVVRIVTATREPEKFGMPDAKAWIAYGASPR
TH_3229|M.thermoresistible__bu LLRLQEVAANNFVHHALVDYVVRVVTATREPEKFGMPDAKAWIAYGASPR
MMAR_2286|M.marinum_M LLRLQDIAANNFVHHALVDYVVRVVMATRQPEQLGMNDVKTWIAFGASPR
MUL_1488|M.ulcerans_Agy99 LLRLQDIAANNFVHHALVDYVVRVVMATRQPEQLGMNDVKTWIAFGASPR
Mb1515|M.bovis_AF2122/97 LLRLQEIAANNFVHHALVDYVVRVVFATRKPEQLGMNDVKSWVAFGASPR
Rv1479|M.tuberculosis_H37Rv LLRLQEIAANNFVHHALVDYVVRVVFATRKPEQLGMNDVKSWVAFGASPR
MLBr_01810|M.leprae_Br4923 LLRLQEISANNFVHHALVDYVVRVVTATRHPEQLGMNDVKTWISFGASPR
MAV_3299|M.avium_104 LVRLQDIAANVFVHHALVDYVVRVVTATRHPEQLGMNDVKTWISFGASPR
MAB_2726c|M.abscessus_ATCC_199 LLRLQDLAANNFVHHALVDYVVRVIAATRNPAQFGLGDVANWISYGASPR
*:***:::** ************:: ***.* ::*: *. *:::*****
Mflv_3661|M.gilvum_PYR-GCK ASLGIIAASRALALVRGRDYVIPQDVVEVIPDVLRHRLVLTYDALADEIS
Mvan_2749|M.vanbaalenii_PYR-1 ASLGIIAASRALALVRGRDYVIPQDVVEIIPDVLRHRLVLTYDALADEIS
MSMEG_3147|M.smegmatis_MC2_155 ASLGIISAARALALVRGRDYVIPQDVVEVIPDVLRHRLVLTYDALADEIS
TH_3229|M.thermoresistible__bu ASLGIIAAARALALVRGRDYVIPQDVVEVIPDVLRHRLVLTYDALADEVT
MMAR_2286|M.marinum_M ASLGIIAASRALALVRGRDYVIPQDIIEVVPDVLRHRLVLTYDALADEIS
MUL_1488|M.ulcerans_Agy99 ASLGIIAASRALALVRGRDYVIPQDIIEVVPDVLRHRLVLTYDALADEIS
Mb1515|M.bovis_AF2122/97 ASLGIIAAARSLALVRGRDYVIPQDVIEVIPDVLRHRLVLTYDALADEIS
Rv1479|M.tuberculosis_H37Rv ASLGIIAAARSLALVRGRDYVIPQDVIEVIPDVLRHRLVLTYDALADEIS
MLBr_01810|M.leprae_Br4923 ASLGIIKASRSLALVRGRDYVIPQDVIEVIPDVLRHRLVLTYNALADEIS
MAV_3299|M.avium_104 ASLGIIAGARSLALVRGRDYVIPQDVVDVIPDVLRHRLVLTYDALADEIS
MAB_2726c|M.abscessus_ATCC_199 ASLGIIASARALALVRGRDYVIPQDVIEVIPDVLRHRLVMSYDALADEVS
****** .:*:**************:::::*********::*:*****::
Mflv_3661|M.gilvum_PYR-GCK AETVVNRILQTVALPQVNAIPQQGHSVPPV------VPAAAAAASGR
Mvan_2749|M.vanbaalenii_PYR-1 SETVVNRILQTVALPQVNAIPQQGHSVPPV------VPAAAAAASGR
MSMEG_3147|M.smegmatis_MC2_155 SETVVNRILQTVALPQVNALPQQGHSVPPA------VPAAAAAASGR
TH_3229|M.thermoresistible__bu AETVINRILQTVALPQVNAVPQQAPPAQP-------MPTAATAASGR
MMAR_2286|M.marinum_M PEIVINRVLQTVALPQVNAVPQQGHSVPP-------VMQAAAAAGGR
MUL_1488|M.ulcerans_Agy99 PEIVINRVLQTVALPQVNAVPQQGHSVPP-------VMQAAAAAGGR
Mb1515|M.bovis_AF2122/97 PEIVINRVLQTVALPQVNAVPQQGHSVPP-------VMQAAAAASGR
Rv1479|M.tuberculosis_H37Rv PEIVINRVLQTVALPQVNAVPQQGHSVPP-------VMQAAAAASGR
MLBr_01810|M.leprae_Br4923 PEIVINRVLQTVALPQVNSFPQQGHSVPS-------VTQAAAAASGR
MAV_3299|M.avium_104 PEIVINRVLQTVALPQVNAVPQQGHSVPP-------VMQPAGAASGR
MAB_2726c|M.abscessus_ATCC_199 ADTAINRILQTVGLPQVNAVPQQGNSVPPAMVGTNNPVPAAHNAAPR
.: .:**:****.*****:.***. .. . .* *. *