For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVLRANGSQPTSTAGAAGDLGAGSDVSGAPGIRDVDGVLEKYRRTRLQRALFDVRGSQAGTAYDEFVDTA GEVRPAWRELAECVSERGRSGLDRLREQVNALVDADGITYIQVDRHGEIVIDREGAAMPGPWHLDALPMV ISAEEWDVLEAGLVQRSRLLDAVLADLYGERRSITSGVLPAQLLFSHPGYLRAAHGITVPGRHQLFLHGC DISRGDDGAFVVNADWTQAPSGAGYALADRRVIAHAVSELYERIAPRPVSPWAQALRLALIDVAPESAEE PVVVVLSPGIHSETAFDQAYLAGVLGFPLVESADLVVRDGKLWMRSLGTLKRVDVVLRRVDADYVDPLDL RPDSRLGVVGLVEVLRRGAVTVVNSLGSGVLESPGVTRFLPQLAQLLLDETPQLDTAPLYWGGIDIERSH LLTRLDSLLIRPVCGGKPIVGPELSTAQREELAARIEATPWQWVGQELPQYSTAPVDFRSSGLSSAGVGM RLFTVAQRSGYAPMVGGLGYLMAPGNAAFRTNTVAAKDIWVRTPDRVTAERIQAPVELATPETRPSPTRV VSSPRVLADLFWLGRYAERAESTARLLNVTRERYHEYRYRQAFEESRCVPVLLGALGSIKGTDTGARDDD HEAIATAPTTLWSVTADRYRPGSLAQSVERLGLAARAVRDQMSNDTWMVLAGVERAVLRTSSIPPDSHSA GETYLANAHSQTLAGMLALSGVVSESMVQDIGWTMMDIGKRIERGLHLTALLRATLAQVHSPAAEHAVAE STLVACESSVIYRRRNPGRISVAAIAELVLFDAENPRSLIYQLERLRTRLKTLPGSSGSSRSERLVDEIA TRLRRLDPDDLETVTDGMRVELADLLDAIHIGLRDLATVITAAHLSVPGGMQPLWGPDERRILP
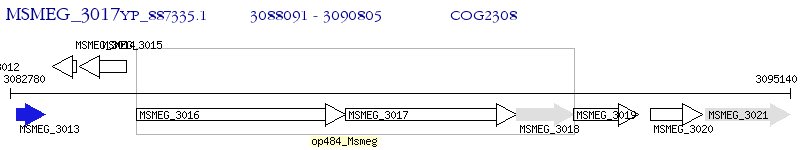
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3017 | - | - | 100% (904) | hypothetical protein MSMEG_3017 |
| M. smegmatis MC2 155 | MSMEG_4570 | - | 7e-66 | 32.37% (519) | hypothetical protein MSMEG_4570 |
| M. smegmatis MC2 155 | MSMEG_4569 | - | 2e-10 | 22.51% (311) | hypothetical protein MSMEG_4569 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2597 | - | 0.0 | 67.61% (883) | hypothetical protein Mb2597 |
| M. gilvum PYR-GCK | Mflv_3770 | - | 0.0 | 72.72% (876) | hypothetical protein Mflv_3770 |
| M. tuberculosis H37Rv | Rv2567 | - | 0.0 | 67.50% (883) | hypothetical protein Rv2567 |
| M. leprae Br4923 | MLBr_02679 | - | 0.0 | 63.16% (874) | hypothetical protein MLBr_02679 |
| M. abscessus ATCC 19977 | MAB_2858c | - | 0.0 | 63.51% (877) | hypothetical protein MAB_2858c |
| M. marinum M | MMAR_2164 | - | 0.0 | 67.61% (886) | hypothetical protein MMAR_2164 |
| M. avium 104 | MAV_3438 | - | 0.0 | 66.25% (889) | hypothetical protein MAV_3438 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1748 | - | 0.0 | 67.49% (886) | hypothetical protein MUL_1748 |
| M. vanbaalenii PYR-1 | Mvan_2629 | - | 0.0 | 74.26% (878) | hypothetical protein Mvan_2629 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3770|M.gilvum_PYR-GCK ------MAVPATPPTPRLGAGRT----------------DVDNPLEPYGS
Mvan_2629|M.vanbaalenii_PYR-1 ------MAHPAPSPTPAPGAGST----------------DVDNPLVPYGT
MSMEG_3017|M.smegmatis_MC2_155 ------MVLRANGSQPTSTAGAAGDLGAGSDVSGAPGIRDVDGVLEKYRR
Mb2597|M.bovis_AF2122/97 -------------MAPSASAATNG--------------YDVDRLLAGYRT
Rv2567|M.tuberculosis_H37Rv -------------MAPSASAATNG--------------YDVDRLLAGYRT
MMAR_2164|M.marinum_M ------------MSRPAAVAAAEP--------------YDVDRLLAGYRT
MUL_1748|M.ulcerans_Agy99 --------MVLGMSSPAAVAAAEP--------------YDVDRLLAGYRT
MAV_3438|M.avium_104 ---MAFADRTFGTGGPSAAAGGS---------------YDADRLLAGYRT
MLBr_02679|M.leprae_Br4923 MIRYMVSTEMALSNSASSSARFNH--------------SDADRLLDGYRA
MAB_2858c|M.abscessus_ATCC_199 ------MAAPASGAPVGPGFAVGD-----------------NPLIAQYAS
: : *
Mflv_3770|M.gilvum_PYR-GCK MRAQPSLFDVASA--FGGLAGYDEFVDTAGDVRPAWQELADCVRDRGRGG
Mvan_2629|M.vanbaalenii_PYR-1 MRAQQVLFEQPQF--GGPTTGYDEFVDAAGNVRPAWQELADCVRDRGRDG
MSMEG_3017|M.smegmatis_MC2_155 TRLQRALFDVRGS--QAGTA-YDEFVDTAGEVRPAWRELAECVSERGRSG
Mb2597|M.bovis_AF2122/97 ARAQETLFDLR----DGPGAGYDEFVDDDGNVRPTWTELADAVAERGKAG
Rv2567|M.tuberculosis_H37Rv ARAQETLFDLR----DGPGAGYDEFVDDDGNVRPTWTELADAVAERGKAG
MMAR_2164|M.marinum_M ARAQEALFDVR----DSPGIGYDEFVDADGNVRPAWTELADAVAERGRTG
MUL_1748|M.ulcerans_Agy99 ARAQEALFDVR----DSPGIGYDEFVDADGNVRPAWTELADAVAERGRTG
MAV_3438|M.avium_104 ARAQQALFDLR----QGPASGYDEFVGPDGKVRPAWTELADAIGERGRAG
MLBr_02679|M.leprae_Br4923 ARAQKVLFDLHSAGTAGPGIGYDEFVDDSGYVRPSWTELADAIGGRGRAG
MAB_2858c|M.abscessus_ATCC_199 ARAQGALFEVGR---RGDQAGYDEFLAPDGSVRPAWTDLADAIAQRGRPG
* * **: . ****: * ***:* :**:.: **: *
Mflv_3770|M.gilvum_PYR-GCK LDRLRAVVRDLVDNDGITYVQTDR--------EGEPVAGPWRLDALPLVI
Mvan_2629|M.vanbaalenii_PYR-1 LDRLRAVVRGLVDNDGITYIQTD---------DGDAVPGPWHLDALPLVI
MSMEG_3017|M.smegmatis_MC2_155 LDRLREQVNALVDADGITYIQVDRHGEIVIDREGAAMPGPWHLDALPMVI
Mb2597|M.bovis_AF2122/97 LDRLRSVVHSLIDHDGITYTAIDAHRDAL-TGDHDLEPGPWRLDPLPLVI
Rv2567|M.tuberculosis_H37Rv LDRLRSVVHSLIDHDGITYTAIDAHRDAL-TGDHDLEPGPWRLDPLPLVI
MMAR_2164|M.marinum_M LDRLRSVVHSLIDHDGITYTGVDSGRED--HGVHGLEPGPWRLDTLPLVV
MUL_1748|M.ulcerans_Agy99 LDRLRSVVHSLIDHDGITYTGVDSGRED--HGVHGLEPGPWRLDTLPLVV
MAV_3438|M.avium_104 LDRLRSVVRGLIDHDGITYTDVEP-------GGRGQEPRPWQLDTLPIVL
MLBr_02679|M.leprae_Br4923 LHQLRSVMHRLIYNDGITYTGIDPG--AL-TNDHGVVPGLWRLDSLPLLV
MAB_2858c|M.abscessus_ATCC_199 LDRLLDRVHTLVDSDGITYMDPRRAPGSVSGPDAPAHPVPWRLDGLPLLL
*.:* :. *: ***** . *:** **:::
Mflv_3770|M.gilvum_PYR-GCK SASDWDHLEAGLVQRSRLLDAVLTDVYGERRVVTGGVLPPELLFSHPGYL
Mvan_2629|M.vanbaalenii_PYR-1 SAADWDRLEAGLVQRSRLLDAVLADLYGERRSITGGVLPPQLLFAHPGYV
MSMEG_3017|M.smegmatis_MC2_155 SAEEWDVLEAGLVQRSRLLDAVLADLYGERRSITSGVLPAQLLFSHPGYL
Mb2597|M.bovis_AF2122/97 SAADWEVLEAGLVQRSRLLDAILADLYGPRSMLTEGVLPPEMLFAHPGYV
Rv2567|M.tuberculosis_H37Rv SAADWEVLEAGLVQRSRLLDAILADLYGPRSMLTEGVLPPEMLFAHPGYV
MMAR_2164|M.marinum_M SAQDWEVLEAGLVQRSRLLDAVLADLYGPRRLLTEGLLPSELVFGHPAYV
MUL_1748|M.ulcerans_Agy99 SARDWEVLEAGLVQRSRLLDAVLADLYGPRRLLTEGLLPSELVFGHPAYV
MAV_3438|M.avium_104 SAADWEPLEAGLLQRSRVLDAVLADLYGPRSLLTEGVLPPELLFGHPGYL
MLBr_02679|M.leprae_Br4923 SATDWEVLEAGLIQRSRLLDAVLTDLYGPRRLLTDGVLPPELLFGHPGYV
MAB_2858c|M.abscessus_ATCC_199 DPADWNVLEVGLTQRATLLDAVLSDLYGAQELISSGALPPQLLFAHPGYV
.. :*: **.** **: :***:*:*:** : :: * **.:::*.**.*:
Mflv_3770|M.gilvum_PYR-GCK RAANGIEIPGRHQLFLHGCDISRNGTGDFVVNADYTQAPSGAGYALADRR
Mvan_2629|M.vanbaalenii_PYR-1 RAARGIEVPGRHQLFLHGCDVSRGSSGDFVVNADWTQAPSGAGYALADRR
MSMEG_3017|M.smegmatis_MC2_155 RAAHGITVPGRHQLFLHGCDISRGDDGAFVVNADWTQAPSGAGYALADRR
Mb2597|M.bovis_AF2122/97 RAANGIQMPGRHQLFMHACDLSRLPDGTFQVNADWTQAPSGSGYAMADRR
Rv2567|M.tuberculosis_H37Rv RAANGIQMPGRHQLFMHACDLSRLPDGTFQVNADWTQAPSGSGYAMADRR
MMAR_2164|M.marinum_M RGANGIEVPGHHQLFLHGCDISRIPDGSYQVNADWTQAPSGAGYALADRR
MUL_1748|M.ulcerans_Agy99 RGANGIEVPGHHQLFLHGCDISRIPDGSYQVNADWTQAPSGAGYALADRR
MAV_3438|M.avium_104 RAANGIEIPGRHQLFMHACDVSRRPDGGFAVNADRTQAPSGAGYALADRR
MLBr_02679|M.leprae_Br4923 RTDHGIEVPGHHQLFMHGCDLSRLPDGGFQINADWTQAPSGAGYALADRR
MAB_2858c|M.abscessus_ATCC_199 RAAHGMALPGRHQLFLLGCDISRTATGDFRVNADWTQAPSGAGYAMADRR
* .*: :**:****: .**:** * : :*** ******:***:****
Mflv_3770|M.gilvum_PYR-GCK VVAHAIPDLYEQIGPRQSSPWAQALRLALVDAAPDAVDEPMVVVLSPGIH
Mvan_2629|M.vanbaalenii_PYR-1 VIAHAIPELYEQIGPQPVSPWAQALRLALVDAAPESAEEPMVVVLSPGIH
MSMEG_3017|M.smegmatis_MC2_155 VIAHAVSELYERIAPRPVSPWAQALRLALIDVAPESAEEPVVVVLSPGIH
Mb2597|M.bovis_AF2122/97 VVAHAVPDLYEELAPRPTTPFAQALRLALIDAAPDVAQDPVVVVLSPGIY
Rv2567|M.tuberculosis_H37Rv VVAHAVPDLYEELAPRPTTPFAQALRLALIDAAPDVAQDPVVVVLSPGIY
MMAR_2164|M.marinum_M VVAHAIPDLYENIAPRPTTPFAQALRLALIDAAPDVAQDPVVVVLSPGIY
MUL_1748|M.ulcerans_Agy99 VVAHAIPDLYENIAPRPTTPFAQALRLALIDAAPDVAQDPVVVVLSPGIY
MAV_3438|M.avium_104 VVAHAIPDLYERIAPRPTTPFAQALRLALIDAAPDVAQDPVVVVLSPGIY
MLBr_02679|M.leprae_Br4923 VVAQALPDLYETIGPRPTTPFAQALRLALIDAAPEVAQDPVVVVLSPGIY
MAB_2858c|M.abscessus_ATCC_199 VVAHAAPDLYEKVAPRPTSSFAQTLRLALIDVAPESVEDPTIVVLSPGIH
*:*:* .:*** :.*: :.:**:*****:*.**: .::* :*******:
Mflv_3770|M.gilvum_PYR-GCK SETAFDQAYLASVLGLPLVESADLVVRDGMLWMRSMGTLKRVDVVLRRVD
Mvan_2629|M.vanbaalenii_PYR-1 SETAFDQAYLASVLGLPLVESADLVVRDGKLWMRSMGTLKRVDVVLRRVD
MSMEG_3017|M.smegmatis_MC2_155 SETAFDQAYLAGVLGFPLVESADLVVRDGKLWMRSLGTLKRVDVVLRRVD
Mb2597|M.bovis_AF2122/97 SETAFDQAYLATLLGFPLVESADLVVRDGKLWMRSLGTLKRVDVVLRRVD
Rv2567|M.tuberculosis_H37Rv SETAFDQAYLATLLGFPLVESADLVVRDGKLWMRSLGTLKRVDVVLRRVD
MMAR_2164|M.marinum_M SETAFDQAYLATLLGFPLVESADLVVRDGKLWMRSLGTLKRVDVVLRRVD
MUL_1748|M.ulcerans_Agy99 SETAFDQAYLATLLGFPLVESADLVVRDGKLWMRSLGTLKRVDVVLRRVD
MAV_3438|M.avium_104 SETAFDQAYLATLLGFPLVESADLVVRDGMVWMRSLGTLKRVDVVLRRVD
MLBr_02679|M.leprae_Br4923 SETAFDQAYLATLLGFQLVESSDLVVCDGKVWMRSLGTLKRVDVVLRRVD
MAB_2858c|M.abscessus_ATCC_199 SETSFDQAYLASVLGFPLVESADLVVRDGKLWMRSLGTLRRVDVVLRRVD
***:******* :**: ****:**** ** :****:***:**********
Mflv_3770|M.gilvum_PYR-GCK ADYADPLDLRPASRLGVVGLVEVLRRGAVTVVNSLGSGVLESPGLLRFLP
Mvan_2629|M.vanbaalenii_PYR-1 ADYADPLDLRPDSRLGVVGLVEVLRRGAVTVVNTLGSGVLESPGLLRFLP
MSMEG_3017|M.smegmatis_MC2_155 ADYVDPLDLRPDSRLGVVGLVEVLRRGAVTVVNSLGSGVLESPGVTRFLP
Mb2597|M.bovis_AF2122/97 AHYADPLDLRADSRLGVVGLVEAQHRGTVTVVNTLGSGILENPGLLRFLP
Rv2567|M.tuberculosis_H37Rv AHYADPLDLRADSRLGVVGLVEAQHRGTVTVVNTLGSGILENPGLLRFLP
MMAR_2164|M.marinum_M AAYADPLDLRADSRLGVVGLVEAQHRGAVTVVNTLGSGILESPGLMRFLP
MUL_1748|M.ulcerans_Agy99 AAYADPLDLRADSRLGVVGLVEAQHRGAVTVVNTLGSGILESPGLMRFLP
MAV_3438|M.avium_104 AEYCDPLDLRADSRLGVAGLVEAQHRGTVTVVNTLGSGILENPGLQRFLP
MLBr_02679|M.leprae_Br4923 ADYMDPLDLRADSLLGVAGLVEAQHRGTVTVVNTLGSGILESAGLLRFLP
MAB_2858c|M.abscessus_ATCC_199 ADYSDPLDLRADSQLGVVGLVEVLRRGAVTVVNSLGSGVLENPGLARFLP
* * ******. * ***.****. :**:*****:****:**..*: ****
Mflv_3770|M.gilvum_PYR-GCK EMAEHLLGETPLLPTPHLYWGGIDIERSHLVSNLGSLVIRPVTGGEPIVG
Mvan_2629|M.vanbaalenii_PYR-1 QLAETLLGETPLLATAPLYWGGSDIERSHLLANLSSLLIRPVNGGEAIVG
MSMEG_3017|M.smegmatis_MC2_155 QLAQLLLDETPQLDTAPLYWGGIDIERSHLLTRLDSLLIRPVCGGKPIVG
Mb2597|M.bovis_AF2122/97 QLSERLLDESPLLHTAPVYWGGIASERSHLLANVSSLLIKSTVSGETLVG
Rv2567|M.tuberculosis_H37Rv QLSERLLDESPLLHTAPVYWGGIASERSHLLANVSSLLIKSTVSGETLVG
MMAR_2164|M.marinum_M GLAERLLGEEPMLQSAPMYWGGIATERSHLLANLSSLLIKSTVGGNTRVG
MUL_1748|M.ulcerans_Agy99 GLAERLLGEEPMLQSAPMYWGGIATERSHLLANLSSLLIKSTVGGKTRVG
MAV_3438|M.avium_104 AMARHLLSETLLLPSAPVYWGGIDTERSHLLANLASLLVKSTVGGKTLVG
MLBr_02679|M.leprae_Br4923 QLSERLLGETLLLPSAPVYWAGIDSERLHLLTNLSSLLIKSTVGGKTIVG
MAB_2858c|M.abscessus_ATCC_199 DLSRRLLDEPLALSGGQSWWGGIDTERSHLLANLASLLVKSTAEEEVYTG
::. **.* * :*.* ** **::.: **:::.. : .*
Mflv_3770|M.gilvum_PYR-GCK ATLSAARREELAARITATPWQWVGQEPPEFSSAPSDFLPGGLSSSSVGMR
Mvan_2629|M.vanbaalenii_PYR-1 PQLSRAQQESLAARITATPWQWVGQELPQFSSAPADHLPGRLSSSSVGMR
MSMEG_3017|M.smegmatis_MC2_155 PELSTAQREELAARIEATPWQWVGQELPQYSTAPVDFRSSGLSSAGVGMR
Mb2597|M.bovis_AF2122/97 PTLSSAQLADLAVRIEAMPWQWVGQELPQFSSAPTNH-AGVLSSAGVGMR
Rv2567|M.tuberculosis_H37Rv PTLSSAQLADLAVRIEAMPWQWVGQELPQFSSAPTNH-AGVLSSAGVGMR
MMAR_2164|M.marinum_M PTLSSAQLAELAARIEAMPWQWVGQELPAFSSAPTDH-AGVLSSAGVGMR
MUL_1748|M.ulcerans_Agy99 PTLSSAQLAELAARIEAMPWQWVGQELPAFSSAPTDH-AGVLSSAGVGMR
MAV_3438|M.avium_104 PALSSRQLAQLAARIETAPWQWIGQELPQFSSAPTDH-AGVLSSAGVGIR
MLBr_02679|M.leprae_Br4923 PSLSSGQLTEMAARIAAKPWQWVGQELPQFSTAPTDH-DGVLSPASIGMR
MAB_2858c|M.abscessus_ATCC_199 PLESAQTLETLRARIEAQPWRWVGQELPELSVAPTYLRSGALTAAPVGMR
. * : .** : **:*:*** * * ** . *:.: :*:*
Mflv_3770|M.gilvum_PYR-GCK LFTVSQRSGYAPMIGGLGYLLAPGADAYRLKSVAAKDVWVRTATRVTAEA
Mvan_2629|M.vanbaalenii_PYR-1 LFTVSQRSGYAPMIGGLGYLAAPGADGYRLKSVAAKDIWVRPPSRVTVEK
MSMEG_3017|M.smegmatis_MC2_155 LFTVAQRSGYAPMVGGLGYLMAPGNAAFRTNTVAAKDIWVRTPDRVTAER
Mb2597|M.bovis_AF2122/97 LFTVAQRSGYAPMIGGLGYVLAPGPAAYTLKTVAAKDIWVRPTERAHAEV
Rv2567|M.tuberculosis_H37Rv LFTVAQRSGYAPMIGGLGYVLAPGPAAYTLKTVAAKDIWVRPTERAHAEV
MMAR_2164|M.marinum_M LFTVAQRSGYSPMIGGLGYVLAPGPAAYTLRSVAAKDIWVRPTERAIVET
MUL_1748|M.ulcerans_Agy99 LFTVAQRSGYSPMIGGLGYVLAPGPAAYTLRSVAAKDIWVRPTERAIVET
MAV_3438|M.avium_104 LFTVAQRGGYAPMIGGIGYLLAAGPAAYTLKSVAAKDVWVRPTERARAEA
MLBr_02679|M.leprae_Br4923 VFTVAQRSGYAPMIGGLGYALASGIDAYSLKKVAAKDIWVLPTERARTEA
MAB_2858c|M.abscessus_ATCC_199 LFTVAQRGGYAPMIGGLGYVAASGYSGSTLNSFAAKDLWVQQPDRDQTER
:***:**.**:**:**:** *.* . ...****:** . * .*
Mflv_3770|M.gilvum_PYR-GCK VP-LPALAT---------------SSPTQEVSSPRVLSDLFWIGRYAERA
Mvan_2629|M.vanbaalenii_PYR-1 TPALPTLAT---------------SSPTQEVSSPRVLSDLFWIGRYAERA
MSMEG_3017|M.smegmatis_MC2_155 IQAPVELATP-----------ETRPSPTRVVSSPRVLADLFWLGRYAERA
Mb2597|M.bovis_AF2122/97 ITVPVLAP------------PAKTGAGTWAVSSPRVLSDLFWMGRYGERA
Rv2567|M.tuberculosis_H37Rv ITVPVLAP------------PAKTGAGTWAVSSPRVLSDLFWMGRYGERA
MMAR_2164|M.marinum_M VAAPVTGPIVQPILEPSALVPIKTGAGTLGVSSPRVLSDLFWMGRYGERA
MUL_1748|M.ulcerans_Agy99 VAAPVTGPIVQPILEPSALVPIKTGAGTLGVSSPRVLSDLFWMGRYGERA
MAV_3438|M.avium_104 VGVPAVEP------------PAKTAAGTWAVSFPRVLSELFWIGRYGERA
MLBr_02679|M.leprae_Br4923 ITHSLLES------------PPMTGTGTLGVSSPRVLSDLFWMGRYGERA
MAB_2858c|M.abscessus_ATCC_199 ALTVAPLDLP-----------GGRSAGSDAVSSPRVLSDLFWLGRYGERA
: : ** ****::***:***.***
Mflv_3770|M.gilvum_PYR-GCK EHTARLLTVTRERYHEYRYRRTMDGSECVPVLLTALGEITG----TDTGA
Mvan_2629|M.vanbaalenii_PYR-1 EHMARLLTVTRERYHEYRYRRTMDGSECVPVLLTALGEITG----TDTGA
MSMEG_3017|M.smegmatis_MC2_155 ESTARLLNVTRERYHEYRYRQAFEESRCVPVLLGALGSIKG----TDTGA
Mb2597|M.bovis_AF2122/97 ENMARLLIVTRERYHVFRHQQDTDESECVPVLMAALGKITGYDTATGAGS
Rv2567|M.tuberculosis_H37Rv ENMARLLIVTRERYHVFRHQQDTDESECVPVLMAALGKITGYDTATGAGS
MMAR_2164|M.marinum_M ENMARLLMVARERYHLFRHHQDTEESECVPVLMAALGNITG----TDTGA
MUL_1748|M.ulcerans_Agy99 ENMARLLMVARERYYLFRHHQDIEESECVPVLMAALGNITG----TDTGA
MAV_3438|M.avium_104 ESMARLLIVTRDRFHVYRHHQHSEESECVPVLMAALGRITGTDTGTGAAA
MLBr_02679|M.leprae_Br4923 ENMARLLIVTREHYHDSRHRQNTEENACLPVLIAALGKITG----TDTGS
MAB_2858c|M.abscessus_ATCC_199 ENLVRLLSVTRERFHEYRYRQFMEASECVPVLLKALGDITG---------
* .*** *:*:::: *::: : . *:***: *** *.*
Mflv_3770|M.gilvum_PYR-GCK AGDYAEMVATAPTTLWSLTADRHRPGSLAQSVERLGLAARAVRDQMSNDT
Mvan_2629|M.vanbaalenii_PYR-1 GGDYTEMVATAPTTLWSLTADRHRAGSLAQSVERLGLTARAVRDQMSNDT
MSMEG_3017|M.smegmatis_MC2_155 RDDDHEAIATAPTTLWSVTADRYRPGSLAQSVERLGLAARAVRDQMSNDT
Mb2597|M.bovis_AF2122/97 AYDRADMIAVAPSTLWSLTVDPDRPGSLVQSVEGLALAARAVRDQLSNDT
Rv2567|M.tuberculosis_H37Rv AYDRADMIAVAPSTLWSLTVDPDRPGSLVQSVEGLALAAQAVRDQLSNDT
MMAR_2164|M.marinum_M ANDHAEMIAVAPSMLWSMTVDLEWPGSLVQSVEGLALAARAVRDQLSNDT
MUL_1748|M.ulcerans_Agy99 ANDHAEMIAVAPSMLWSMTVDLEWPGSLVQSVEGLALAARAVRDQLSNDT
MAV_3438|M.avium_104 GGDAAETIAVAPSTLWSLTVDPQRPGSLVQSVEGLALAARAVRDQMSNDT
MLBr_02679|M.leprae_Br4923 G-GQADVIAVAPSTLWSLTTDRNLPGSLVQSVDGLVLAARAVRAQLSRDT
MAB_2858c|M.abscessus_ATCC_199 ---TDTAAEDLGTQLWSLTVDKERSGSLAQSVERLGLAARAVRDQMSNDT
: ***:*.* .***.***: * *:*:*** *:*.**
Mflv_3770|M.gilvum_PYR-GCK WMVLSALDRALLNAPDTPPDSQAEGDA-------FLASTNTLALAGMMAL
Mvan_2629|M.vanbaalenii_PYR-1 WMVLSALERTLLNAPGVPPDSQAEGDA-------FLARASTLTLGGMLAL
MSMEG_3017|M.smegmatis_MC2_155 WMVLAGVERAVLRTSSIPPDSHSAGET-------YLANAHSQTLAGMLAL
Mb2597|M.bovis_AF2122/97 WMVLANVERAVEHKS-DPPQSLAEADA-------VLASAQAETLAGMLTL
Rv2567|M.tuberculosis_H37Rv WMVLANVERAVEHKS-DPPQSLAEADA-------VLASAQAETLAGMLTL
MMAR_2164|M.marinum_M WIVLADVERAVTLRS-DPPQSLAEADG-------LLAEAQAQTLAGMLTL
MUL_1748|M.ulcerans_Agy99 WIVLADVERAVTLRS-DPPQSLAEADG-------LLAEAQAQTLAGMLTL
MAV_3438|M.avium_104 WMVLAGVERALALDS-EPPDSLAEADA-------LLTAAQTQTLAGMLTL
MLBr_02679|M.leprae_Br4923 WMVLSEVERAMVHET-EPPESLVEGDA-------LLASVQARTLAGMLTL
MAB_2858c|M.abscessus_ATCC_199 WMVLGGMDRAIAAAAAEYDEFRRSGDAPTRADDTVLAATQTQVLAGLLAL
*:**. ::*:: . : .: *: . : .*.*:::*
Mflv_3770|M.gilvum_PYR-GCK SGVAAESMVHDVGWTMMDIGKRIERGLALTALLRATLTTVRTPGAEQTVT
Mvan_2629|M.vanbaalenii_PYR-1 SGVVAESMVHDVGWMMMDTGKRIERALALTALLRATLTTVRTPGAERTVT
MSMEG_3017|M.smegmatis_MC2_155 SGVVSESMVQDIGWTMMDIGKRIERGLHLTALLRATLAQVHSPAAEHAVA
Mb2597|M.bovis_AF2122/97 SGVAGESMVHDVGWTMMDIGKRIERGLWLTALLQATLSTVRHPAAEQAII
Rv2567|M.tuberculosis_H37Rv SGVAGESMVHDVGWTMMDIGKRIERGLWLTALLQATLSTVRHPAAEQAII
MMAR_2164|M.marinum_M SGVASESMVRDVGWTMMDIGKRIERGLWLTALLADTLTTVRGVVAEQTII
MUL_1748|M.ulcerans_Agy99 SGVASESMVRDVGWTMMDIGKRIERGLWLTALLADTLTTVRGVVAEQTII
MAV_3438|M.avium_104 SGVAGESMVRDVGWTMMDIGKRIERGLWLTALLRATLTVVRGAAAEQTIL
MLBr_02679|M.leprae_Br4923 SGLAGESMVRDIGWTIMDIGKRIERGLALTALLHATLTDVRSPAAEHSII
MAB_2858c|M.abscessus_ATCC_199 SGLAAESTVHDIGWTIMDIGKRIERGLGLTALLRSTLTTERGRAAEQSVT
**:..** *:*:** :** ******.* ***** **: : **:::
Mflv_3770|M.gilvum_PYR-GCK ESALVACESLVIYRRRNPGKISVAGVADLMLFDADNPRSLAYQLERLRTH
Mvan_2629|M.vanbaalenii_PYR-1 ESTLVACESLVIYRRRNPGKISVAGLTELVLFDAGNPRSLVYQLERLRSH
MSMEG_3017|M.smegmatis_MC2_155 ESTLVACESSVIYRRRNPGRISVAAIAELVLFDAENPRSLIYQLERLRTR
Mb2597|M.bovis_AF2122/97 EATLVACESSVIYRRRTVGKFSVAAVTELMLFDAQNPRSLVYQLERLRAD
Rv2567|M.tuberculosis_H37Rv EATLVACESSVIYRRRTVGKFSVAAVTELMLFDAQNPRSLVYQLERLRAD
MMAR_2164|M.marinum_M ESTLEVCESSVIYRRRTVGKFSVTAVNELMLFDAHNPRSLVYQMERLRAN
MUL_1748|M.ulcerans_Agy99 ESTLEVCESSVIYRRRTVGKFRVTAVNELMLFDAHNPRSLIYQLERLRAN
MAV_3438|M.avium_104 ESTLVACESSVSYRRRTAGKVSVAAMAELMLFDAQNPRSLLYQVERLRAD
MLBr_02679|M.leprae_Br4923 ESILVICESSVIYQRRTVSTIDVAVVAELMLFDATNPRSLVYQLDRLRVD
MAB_2858c|M.abscessus_ATCC_199 ESTLVACESSVMYRRRTRGKISLEAVADLLLFDADNPRSLLYQVESLRTD
*: * *** * *:**. . . : : :*:**** ***** **:: **
Mflv_3770|M.gilvum_PYR-GCK LRSLPESTGSSRPERMVDEIAARLRRMEPGELEEVAADGTRDGLRALLTA
Mvan_2629|M.vanbaalenii_PYR-1 LKSLPGSSGSSRPERMVDEIAARLRRIEPAELEAVTADGRRAEFDGLLSG
MSMEG_3017|M.smegmatis_MC2_155 LKTLPGSSGSSRSERLVDEIATRLRRLDPDDLETVT-DGMRVELADLLDA
Mb2597|M.bovis_AF2122/97 LKDLPGSSGSSRPERMVDEMNTRLRRSHPEELEEVSADGLRAELAELLAG
Rv2567|M.tuberculosis_H37Rv LKDLPGSSGSSRPERMVDEMNTRLRRSHPEELEEVSADGLRAELAELLAG
MMAR_2164|M.marinum_M LKDLPGTSGSSRPERMVDEISARLRRSHPAELEEIT-DGRRTELAELLNG
MUL_1748|M.ulcerans_Agy99 LEDLPGTSGSSRPERMVGEISARLRRSHPAELEEIT-DGRRTELAELLNG
MAV_3438|M.avium_104 LKDLPGASGSSRPERLVDEIGTRLRRSHPAELERISDDGRRTELAELLDS
MLBr_02679|M.leprae_Br4923 LADLPGSSRSSLPERMVAEMSTRLRRSAPAQLKEVSADGRRYVLAELLAA
MAB_2858c|M.abscessus_ATCC_199 LKSLPSASGTSRPERLVEDLVTQLRRIDPADLETTGEQGLRDTLAETLDS
* ** :: :* .**:* :: ::*** * :*: :* * : * .
Mflv_3770|M.gilvum_PYR-GCK IHRDLRELSAVITTVHLSLPGDMQPLWGPDERRVVP-
Mvan_2629|M.vanbaalenii_PYR-1 IHHDLRELSSAITAVHLSLPGDMQPLWGPDERRVVP-
MSMEG_3017|M.smegmatis_MC2_155 IHIGLRDLATVITAAHLSVPGGMQPLWGPDERRILP-
Mb2597|M.bovis_AF2122/97 IHASLRDVADVLTATQLALPGGMQPLWGPDQRRVMPA
Rv2567|M.tuberculosis_H37Rv IHASLRDVADVLTATQLALPGGMQPLWGPDQRRVMPA
MMAR_2164|M.marinum_M IHVSLRELAEVITDTQLALPGGMQPLWGPEERRTMPA
MUL_1748|M.ulcerans_Agy99 IHVSLRELAEVITDTQLALPGGMQPLWGPEERRTMPA
MAV_3438|M.avium_104 VHAELRSLAEVLTTTQLALPGGMQPLWGPDVRRVMPA
MLBr_02679|M.leprae_Br4923 VDTALRDLADAITATQLAVPCGMQPLWGPDNRQTMS-
MAB_2858c|M.abscessus_ATCC_199 VHHALRELSSVVTATHLSMPTGMQPLWGPDTRRRLP-
:. **.:: .:* .:*::* .*******: *: :.