For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LARNAESLYWIGRYVERADDTARILDVTVHQLLEDSSVDPDQASRTLLRVLGIDPPRERLDVWSLTDIVA FSRGTYGGASIVDAISAARENARGAREVTSTEIWECLNTTYNALAERERAAKRLGPHEFLSYVEGRAAMF AGLADSTLSRDDGYRFMVLGRAIERVDMTVRLLLSRVGDSASSPAWVTLLRSAGAHDTYLRTYRGVLDAG RVVEFMMLDRLFPRSIFHSLRLAEHNLDELLNRPHNRLGATDEAQRLLGRARSELEFLQPGALLESLETR LAGLQKTCRDVGEALAQQYFHSAPWVAWTDAGRGDALVIEEGEI*
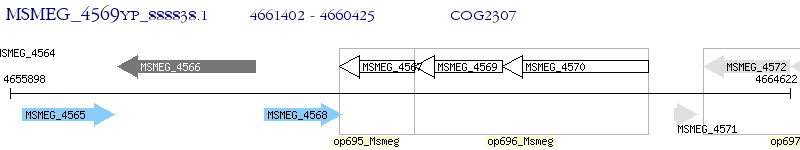
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4569 | - | - | 100% (325) | hypothetical protein MSMEG_4569 |
| M. smegmatis MC2 155 | MSMEG_3017 | - | 6e-11 | 22.51% (311) | hypothetical protein MSMEG_3017 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2433c | - | 1e-158 | 84.88% (324) | hypothetical protein Mb2433c |
| M. gilvum PYR-GCK | Mflv_2667 | - | 1e-171 | 93.85% (325) | hypothetical protein Mflv_2667 |
| M. tuberculosis H37Rv | Rv2410c | - | 1e-158 | 84.88% (324) | hypothetical protein Rv2410c |
| M. leprae Br4923 | MLBr_00606 | - | 1e-154 | 83.02% (324) | hypothetical protein MLBr_00606 |
| M. abscessus ATCC 19977 | MAB_1636 | - | 1e-153 | 83.44% (326) | hypothetical protein MAB_1636 |
| M. marinum M | MMAR_3731 | - | 1e-155 | 82.72% (324) | hypothetical protein MMAR_3731 |
| M. avium 104 | MAV_1772 | - | 1e-154 | 82.72% (324) | hypothetical protein MAV_1772 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3674 | - | 1e-155 | 82.41% (324) | hypothetical protein MUL_3674 |
| M. vanbaalenii PYR-1 | Mvan_3910 | - | 1e-169 | 92.62% (325) | hypothetical protein Mvan_3910 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2667|M.gilvum_PYR-GCK MLARNAESLYWIGRYVERADDTARILDVTVHQLLEDSSVDPDQASRTLLK
Mvan_3910|M.vanbaalenii_PYR-1 MLARNAESLYWIGRYVERADDTARILDVTVHQLLEESSVDPDQASRTLLK
MSMEG_4569|M.smegmatis_MC2_155 MLARNAESLYWIGRYVERADDTARILDVTVHQLLEDSSVDPDQASRTLLR
MMAR_3731|M.marinum_M MLARNAEALYWIGRYVERADDTARILDVAIHQLLEDSSIDPDHASRLLLR
MUL_3674|M.ulcerans_Agy99 MLARNAEALYWIGRYVERADDTARILDVAIHQLLEDSSIDPDHASRLLLR
Mb2433c|M.bovis_AF2122/97 MLARNAEALYWIGRYVERADDTARILDVAVHQLLEDSSVDPDQASRLLLR
Rv2410c|M.tuberculosis_H37Rv MLARNAEALYWIGRYVERADDTARILDVAVHQLLEDSSVDPDQASRLLLR
MLBr_00606|M.leprae_Br4923 MLARNAEALYWIGRYVERADDTARILDVVVHQLLEDSSVDPDHASRTLLR
MAV_1772|M.avium_104 MLARNAEALYWIGRYVERADDTARILDVALHQLLEDSSVDPDQASRVLLR
MAB_1636|M.abscessus_ATCC_1997 MLARNAESLYWIGRYVERADDTARILDVTVHQLLEDASVDPDHASRVLLR
*******:********************.:*****::*:***:*** **:
Mflv_2667|M.gilvum_PYR-GCK VLGIDPPDGALDVWSLTDIVAFSRDSVGACSIVDAISAARENARGAREVT
Mvan_3910|M.vanbaalenii_PYR-1 VLGIDPPDGQLDVWSLTDIVAFSRETYGGCSIVEAISAARENARGAREVT
MSMEG_4569|M.smegmatis_MC2_155 VLGIDPPRERLDVWSLTDIVAFSRGTYGGASIVDAISAARENARGAREVT
MMAR_3731|M.marinum_M VLGIDPPDHELDVWSLTDLVAFSTNASGGCSIVEAISAARENAKSAREVT
MUL_3674|M.ulcerans_Agy99 VLGIDPPDHELDVWSLTDLVAFSTNASGGCSIVEAISAARENAKSAREVT
Mb2433c|M.bovis_AF2122/97 VLGIEPPDHELDVWSLTDLVAFSTNSQGGSSIVDAISAARENAKSAREVT
Rv2410c|M.tuberculosis_H37Rv VLGIEPPDHELDVWSLTDLVAFSTNSQGGSSIVDAISAARENAKSAREVT
MLBr_00606|M.leprae_Br4923 VLGIELPDHELDVWSLTDLVAFSTNAQGGSSIVDAISAARENAKSAREVT
MAV_1772|M.avium_104 VLGIDPPDHDLDVWSLTDLVAYSTGAQGGCSIVDAVTAARENAKSAREVT
MAB_1636|M.abscessus_ATCC_1997 VLGIEAPDTVLDVWSLTELVAFSRGVQGG-SIVDSISAARENARGAREVT
****: * *******::**:* *. ***::::******:.*****
Mflv_2667|M.gilvum_PYR-GCK STEIWECLNTTYNALAERERAAKRLGPHEFLSYVEGRAAMFAGLADSTLS
Mvan_3910|M.vanbaalenii_PYR-1 SSEIWECLNTTYNALAERERAAKRLGPHEFLSYVEGRAAMFAGLADSTLS
MSMEG_4569|M.smegmatis_MC2_155 STEIWECLNTTYNALAERERAAKRLGPHEFLSYVEGRAAMFAGLADSTLS
MMAR_3731|M.marinum_M SIETWECLNTTYHALPERERAAKRLGPHEFLSFVEGRAAMFAGLADSTLS
MUL_3674|M.ulcerans_Agy99 SIETWECLNTTYHALPERERAAKRLGPHEFLSFVEGRAAMFAGLADSTLS
Mb2433c|M.bovis_AF2122/97 SSETWECLNTTYNALPERERAAKRLGPHEFLSFIEGRAAMFAGLADSTLL
Rv2410c|M.tuberculosis_H37Rv SSETWECLNTTYNALPERERAAKRLGPHEFLSFIEGRAAMFAGLADSTLL
MLBr_00606|M.leprae_Br4923 SSETWECLNTTYHALAERERAAKRLGPHEFLSFIEGRAAMFAGLADSTLS
MAV_1772|M.avium_104 SSEIWECLNTTYHALPERERAAKRLGPHDFLSFIERRAAMFAGLADSTLS
MAB_1636|M.abscessus_ATCC_1997 STEMWECLNTTFNALPDRERAARRLGPHEFFTYVEGRAAQFAGLADSTLS
* * *******::**.:*****:*****:*::::* *** *********
Mflv_2667|M.gilvum_PYR-GCK RDDGYRFMVLGRALERVDMTVRLLLSRVGDSASSPAWVTLLRSAGAHDTY
Mvan_3910|M.vanbaalenii_PYR-1 RDDGYRFMVLGRALERVDMTVRLLLSRVGDSASSPAWVTLLRSAGAHDTY
MSMEG_4569|M.smegmatis_MC2_155 RDDGYRFMVLGRAIERVDMTVRLLLSRVGDSASSPAWVTLLRSAGAHDTY
MMAR_3731|M.marinum_M RDDGYRFMLLGRAIERVDMTVRLLLSRVGDSASSPAWVTLLRSVGAHDTY
MUL_3674|M.ulcerans_Agy99 RDDGYRFMLLGRAIERVDMTVRLLLSRVGDSASSPAWATLLRSVGAHDTY
Mb2433c|M.bovis_AF2122/97 RDDGYRFMLLGRAIERVDMTVRLLLSRVGDSASSPAWVTLLRSAGAHDTY
Rv2410c|M.tuberculosis_H37Rv RDDGYRFMLLGRAIERVDMTVRLLLSRVGDSASSPAWVTLLRSAGAHDTY
MLBr_00606|M.leprae_Br4923 RDDGYRFMLLGRAIERVDMTVRLLLSRVGDSASSPAWVTLLRSAGGHDTY
MAV_1772|M.avium_104 RDDGYRFMVLGRAIERVDMTVRLLLSRVGDSASSPAWVTLLRSAGAHDTY
MAB_1636|M.abscessus_ATCC_1997 RDDGYRFLVLGRSIERVDMTVRLLLSRVGDRASSPAWVTLLRSAGAHDTY
*******::***::**************** ******.*****.*.****
Mflv_2667|M.gilvum_PYR-GCK LRTYRGVLDAGRVVEFMMLDRLFPRSIFYSLRLAEHSLDELMNRPHNRLG
Mvan_3910|M.vanbaalenii_PYR-1 LRTYRGVLDAGRVVEFMLLDRLFPRSIFYSLRLAEHSLDELMNRPHNRLG
MSMEG_4569|M.smegmatis_MC2_155 LRTYRGVLDAGRVVEFMMLDRLFPRSIFHSLRLAEHNLDELLNRPHNRLG
MMAR_3731|M.marinum_M LRTYRGVLDAARVVEFMMLDRLFPRSVYYSLRMAEHNLDELLHNPQSRIG
MUL_3674|M.ulcerans_Agy99 LHTYRGVLDAARVVEFMMLDRLFPRSVYYSLRMAEHNLDELLHNPHSRIG
Mb2433c|M.bovis_AF2122/97 LRTYRGVLDAGRVVEFMMLDRLFPRSVFHSLKLAEHNLAELMHNPHSRIG
Rv2410c|M.tuberculosis_H37Rv LRTYRGVLDAGRVVEFMMLDRLFPRSVFHSLKLAEHNLAELMHNPHSRIG
MLBr_00606|M.leprae_Br4923 LRTYRGVLDAARVVEFMLLDRLFPRSIFYSLKLAEHNLDELLRNPQSRVG
MAV_1772|M.avium_104 LRTYRGVLDAGRVVEFMLLYRLFPRSVFYSLRLAEHNLDELMHNRQSRVG
MAB_1636|M.abscessus_ATCC_1997 LRTYRGVLDANRVVEFMLLDRLFPRSVFHSLRQAEQSLDELDHRPHSRVG
*:******** ******:* ******:::**: **:.* ** .. :.*:*
Mflv_2667|M.gilvum_PYR-GCK ATAEAQRLLGRARSELEFLQPGALLDSLETRLADLQKTCFDVGEALALQY
Mvan_3910|M.vanbaalenii_PYR-1 TTAEAQRLLGRARSELEFLQPGVLLDSLEERLAGLQKTIFDVGEALALQY
MSMEG_4569|M.smegmatis_MC2_155 ATDEAQRLLGRARSELEFLQPGALLESLETRLAGLQKTCRDVGEALAQQY
MMAR_3731|M.marinum_M ATTEAQRLLGQARSKLEFVQPGVLLESLDSRLSGLQRTCRDVGEALTLQY
MUL_3674|M.ulcerans_Agy99 ATTEAQRLLGQARSKLEFVQPGVLLESLDSRLSGLQRTCRDVGEALTLQY
Mb2433c|M.bovis_AF2122/97 ATTEAQRLLGQARSELEFVQPGVLLETLESRLAGLQTTCRDVGDALALQY
Rv2410c|M.tuberculosis_H37Rv ATTEAQRLLGQARSELEFVQPGVLLETLESRLAGLQTTCRDVGDALALQY
MLBr_00606|M.leprae_Br4923 ATTEAQRLLGQARSELEFVQPGVLLKTLDSRLASLQTTCRDVGDALVLQY
MAV_1772|M.avium_104 ATAEAQRLLGQARSELEFVQPGVLLESLESRLAGLQRTCRDVSDALALQY
MAB_1636|M.abscessus_ATCC_1997 ARAEAQRLLGRARSELEFMRPGVLLEDLPTRLAGLQQTCRELGEAVALQY
: *******:***:***::**.**. * **:.** * ::.:*:. **
Mflv_2667|M.gilvum_PYR-GCK FHSAPWVAWTDAGR-NALVIEE-GEI
Mvan_3910|M.vanbaalenii_PYR-1 FHSAPWVAWTDAGR-NALVIEE-GEI
MSMEG_4569|M.smegmatis_MC2_155 FHSAPWVAWTDAGRGDALVIEE-GEI
MMAR_3731|M.marinum_M FHVTPWVAWTDAGPRGQMVARK-GES
MUL_3674|M.ulcerans_Agy99 FHVTPWVAWTDAGPRGQMVARK-GES
Mb2433c|M.bovis_AF2122/97 FHAAPWVAWSDAGQRGQLVGSQ-EES
Rv2410c|M.tuberculosis_H37Rv FHAAPWVAWSDAGQRGQLVGSQ-EES
MLBr_00606|M.leprae_Br4923 FHVAPWVAWSDAGHRGRMVGKR-GEA
MAV_1772|M.avium_104 FHVTPWVAWSDASQRARLVSRK-GDG
MAB_1636|M.abscessus_ATCC_1997 FHAAPWVAWSDAGGRHDVDPIEEGEI
** :*****:**. : . :