For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDPALSGDPVATQTSGVRILLDGVTKRYDAKGPAAVDDITIEFPAGEIVMLVGPSGCGKTTTMKMINRLI EPTSGRILIGDDDVTHRDPDELRRHIGYVIQGAGLFPHYTVGENIAIVPRLLKWEKQRIAARVDELLDLV NLEPAQYRDRYPRELSGGQQQRVGVARALAADPPVLLMDEPFGAVDPITRQHLQDELLRLQDELHKTIVF VTHDFDEAVKVGDRIAILTEGSRIVQYDKPEEILANPADDFVRGFVGHGAALKQLTLTRVRNVELHEAAV AHVGDDPADAIRTAQAIDREHVIVLDQHDRPLRWLSLSELSDAHSLERVTRDENLEKVSTSSTLNDALDV MLTSSHGVVVVTGRRNAYQGVIRVETIMDAMPDMQTSPTGPVL*
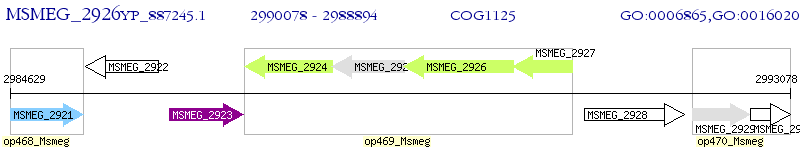
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2926 | - | - | 100% (394) | glycine betaine/carnitine/choline transport ATP-binding protein opuCA |
| M. smegmatis MC2 155 | MSMEG_6333 | - | 5e-80 | 43.58% (374) | amino acid ABC transporter, ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_4530 | cysA | 2e-51 | 43.00% (293) | sulfate ABC transporter, ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3784c | proV | 9e-74 | 43.94% (371) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_3109 | - | 1e-123 | 61.31% (367) | glycine betaine/L-proline ABC transporter, ATPase subunit |
| M. tuberculosis H37Rv | Rv3758c | proV | 9e-74 | 43.94% (371) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | MLBr_01424 | - | 7e-45 | 39.75% (239) | putative binding-protein-dependent transport protein |
| M. abscessus ATCC 19977 | MAB_1226 | - | 1e-178 | 81.96% (377) | ABC transporter ATP-binding protein |
| M. marinum M | MMAR_0034 | proV_1 | 1e-170 | 78.13% (375) | osmoprotectant (glycine |
| M. avium 104 | MAV_0318 | - | 7e-71 | 41.91% (377) | amino acid ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_4520 | - | 1e-174 | 82.34% (368) | PUTATIVE osmoprotectant (glycine |
| M. ulcerans Agy99 | MUL_0033 | proV_1 | 1e-167 | 76.53% (375) | osmoprotectant (glycine |
| M. vanbaalenii PYR-1 | Mvan_5600 | - | 1e-76 | 45.23% (367) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3784c|M.bovis_AF2122/97 ---------------------MICFDDVSKVYAHG-ATAVDRLTLEVPNG
Rv3758c|M.tuberculosis_H37Rv ---------------------MICFDDVSKVYAHG-ATAVDRLTLEVPNG
MAV_0318|M.avium_104 ---------------------MIVFDNVSKVFADG-TAAVDRLSLTVPTG
Mvan_5600|M.vanbaalenii_PYR-1 ---------------------MISFEHVTKRYPDG-TVAVDDLTLEVPEG
MSMEG_2926|M.smegmatis_MC2_155 ---MSDPALSGDPVATQTSGVRILLDGVTKRYDAKGPAAVDDITIEFPAG
MAB_1226|M.abscessus_ATCC_1997 ---MSDVTEN----ATITGGARILLDGVTKLYDGKSTPAVDNVTMEIPAG
MMAR_0034|M.marinum_M MPEAASADQPTTSDAPQSSGLRILLDGVTKRYGVRSTPAVDNVTLEIGAG
MUL_0033|M.ulcerans_Agy99 MPEAASADQPTTSDAPQSSGLRILLDGVTKRYGVRSTPAVDNVTLEIGAG
TH_4520|M.thermoresistible__bu ------VSEP-TSDLQSGGGARILLDGVTKRYDPKSPPAVDNITMEIPAG
Mflv_3109|M.gilvum_PYR-GCK ---MTEQAAP--------TGVRIELDHVSKVYPGAKQPAVDDVSLEIPAG
MLBr_01424|M.leprae_Br4923 -------------------MASVTFEQATRQFPDTDRPALDRLDLDVDDG
: :: .:: : *:* : : . *
Mb3784c|M.bovis_AF2122/97 MLTVFVGPSGCGKTTALRMINRMVDPTSGTITVDGTDVSTVNAVKLRLGI
Rv3758c|M.tuberculosis_H37Rv MLTVFVGPSGCGKTTALRMINRMVDPTSGTITVDGTDVSTVNAVKLRLGI
MAV_0318|M.avium_104 KLTVFVGSSGSGKTTALRMINRMIEPTSGTISIDGVDVSTLDPVKLRLGI
Mvan_5600|M.vanbaalenii_PYR-1 TLAVFVGPSGCGKTTSMRMINRMIEPTSGTLTVDGRDVTKVDAVKLRLGI
MSMEG_2926|M.smegmatis_MC2_155 EIVMLVGPSGCGKTTTMKMINRLIEPTSGRILIGDDDVTHRDPDELRRHI
MAB_1226|M.abscessus_ATCC_1997 ETVMFVGPSGCGKTTTMKMINRLIEPTSGRIFIGDEDVTDKNPDELRRHI
MMAR_0034|M.marinum_M EIVMLVGPSGCGKTTTMKMINRLIEPTSGRIFIGDDDVTRRNPDELRRHI
MUL_0033|M.ulcerans_Agy99 EIVMLVGPSGCGKTTTMKVINRLIEPTSGRIFIGDDDVTRRNPDELRRHI
TH_4520|M.thermoresistible__bu EIVMLVGPSGCGKTTTMKMINRLIEPTSGRIFIGDDDVTERDADELRRHI
Mflv_3109|M.gilvum_PYR-GCK EIVIFVGPSGCGKTTTMRMINRLSEPTSGRILIGDKDALSIKPTELRRSI
MLBr_01424|M.leprae_Br4923 EFVVVVGPSGCGKTTSLRMVAGLETLDSGCIRIGDRDVTHVDPK--DRDV
.:.**.**.****::::: : ** : :.. *. .. :
Mb3784c|M.bovis_AF2122/97 GYVIQNAGLMPHQRVIDNVATVPVLKGQPRRAARKAGYEVLERVGLDP-K
Rv3758c|M.tuberculosis_H37Rv GYVIQNAGLMPHQRVIDNVATVPVLKGQPRRAARKAGYEVLERVGLDP-K
MAV_0318|M.avium_104 GYVIQHAGLMPHQRVIDNVATVPVLRGQRRRAARTAAYQVLERVGLDV-K
Mvan_5600|M.vanbaalenii_PYR-1 GYVIQSAGLMPHLRVVDNVATVPVLRGESRRSARRSALGVLERVGLDP-K
MSMEG_2926|M.smegmatis_MC2_155 GYVIQGAGLFPHYTVGENIAIVPRLLKWEKQRIAARVDELLDLVNLEPAQ
MAB_1226|M.abscessus_ATCC_1997 GYVIQGAGLFPHFTVGDNIAIVPKLLNWDKQRIDERVDELLDLVNLDPAQ
MMAR_0034|M.marinum_M GYVVQGAGLFPHLTVGDNIGIVPRLLKWDKNRVAARIDELLDLVNLDPAQ
MUL_0033|M.ulcerans_Agy99 GYVVQGAGVFLHLTVGDNIGIVPRLLKWDKNRVAARIDELLDLVNLDPAQ
TH_4520|M.thermoresistible__bu GYVIQGAGLFPHYTVAENIAIVPRLLKWDRKRIAARVDELLDLVSLDPDE
Mflv_3109|M.gilvum_PYR-GCK GYAIQQAGLFPHMTIRQNVGLVPGLLGWDRKRIGDRVDELLDLVGLEPAQ
MLBr_01424|M.leprae_Br4923 AMVFQNYALYPHMTVAQNLGFALKISKISKAEIHDRILVVAKLLDLQP--
. ..* .: * : :*:. . : : : . :.*:
Mb3784c|M.bovis_AF2122/97 VATRYPAQLSGGEQQRVGVARALAADPPILLMDEPFSAVDPVVRHELQNE
Rv3758c|M.tuberculosis_H37Rv VATRYPAQLSGGEQQRVGVARALAADPPILLMDEPFSAVDPVVRHELQNE
MAV_0318|M.avium_104 LARRYPAQLSGGEQQRVGVARALAADPPILLMDEPFSAVDPVVRHDLQNE
Mvan_5600|M.vanbaalenii_PYR-1 LADRYPAQLSGGQQQRVGVARALAADPPILLMDEPFSAVDPVVREDLQTE
MSMEG_2926|M.smegmatis_MC2_155 YRDRYPRELSGGQQQRVGVARALAADPPVLLMDEPFGAVDPITRQHLQDE
MAB_1226|M.abscessus_ATCC_1997 YRNRYPRELSGGQQQRVGVARALAADPPVLLMDEPFGAVDPITRQRLQDE
MMAR_0034|M.marinum_M YRDRFPRELSGGQQQRVGVARALAADPPVLLMDEPFGAVDPITRQRLQDE
MUL_0033|M.ulcerans_Agy99 YRDRFPRELSGGQQQRVGVARALAADPPVLLMDEPFGAVDPITRQRLQDE
TH_4520|M.thermoresistible__bu YRDRYPRELSGGQQQRVGVARALAADPPVLLMDEPFGAVDPITRQRLQDE
Mflv_3109|M.gilvum_PYR-GCK YAERYPRQLSGGQQQRVGVARALAADPPVLLMDEPFGAVDPITRSTLQDE
MLBr_01424|M.leprae_Br4923 YLDRKPKDLSGGQRQRVAMGRAIVRRPQVFLMDEPLSNLDAKLRAQTRNQ
* * :****::***.:.**:. * ::*****:. :*. * : :
Mb3784c|M.bovis_AF2122/97 ILRLQAELHKTIVFVTHDIDEALKLADLVAVFAPGGALAQYDETARLLSS
Rv3758c|M.tuberculosis_H37Rv ILRLQAELHKTIVFVTHDIDEALKLADLVAVFAPGGALAQYDETARLLSS
MAV_0318|M.avium_104 ILRLQSELHKTIVFVTHDIDEALRLGERVAVFK-GGMLQQCDEPARLLSR
Mvan_5600|M.vanbaalenii_PYR-1 ILRLQSELRKTIVFVTHDIDEAIKLGDKVAVFGRGGVLQQYDAPARLLSN
MSMEG_2926|M.smegmatis_MC2_155 LLRLQDELHKTIVFVTHDFDEAVKVGDRIAILTEGSRIVQYDKPEEILAN
MAB_1226|M.abscessus_ATCC_1997 LLRLQEELSKTIVFVTHDFDEAVKLGDRIAILREGSQIVQYDTPEVILAN
MMAR_0034|M.marinum_M LLRLQEELHKTIVFVTHDFGEAVKLGDRIAILTTGSKVVQYDTPERILAE
MUL_0033|M.ulcerans_Agy99 LLRLQEELHKTIVFVTHDFGEAVKLGDRIAILTTGSKVVQYDTPERVLAE
TH_4520|M.thermoresistible__bu LLRLQDELRKTIVFVTHDFDEAVKLGDRIAILQQGSKVVQYDTPEEILTN
Mflv_3109|M.gilvum_PYR-GCK LLRLQNELRKSIVFVTHDFGEAVKLGDRIAVLGQQSKVLQYDTPQNILAS
MLBr_01424|M.leprae_Br4923 IAELQCQLGTTTVYVTHDQVEAMTMGDRVAVLCYG-VLQQFATPRELYHK
: .** :* .: *:**** **: :.: :*:: : * . :
Mb3784c|M.bovis_AF2122/97 PANDFVSKFIGLGRGYRWLQLFDAAGLPVRD--IEQVSVNGLSDARDRQV
Rv3758c|M.tuberculosis_H37Rv PANDFVSKFIGLGRGYRWLQLFDAAGLPVRD--IEQVSVNGLSDARDRQV
MAV_0318|M.avium_104 PANDFVSRFIGLGRGYRWLQLIDAAGLPVHD--VRRLFAENLSGA---DV
Mvan_5600|M.vanbaalenii_PYR-1 PANDEVAEFVGADRGYRGLQFFHATGLPLHD--IRRVAEPEINAL---TL
MSMEG_2926|M.smegmatis_MC2_155 PADDFVRGFVGHGAALKQLTLTRVRNVELHEAAVAHVGDDPADAIRTAQA
MAB_1226|M.abscessus_ATCC_1997 PANDFVRGFVGHGAALKQLTMTRVRDVELHEAAVAHVGDDPAQAIEAAES
MMAR_0034|M.marinum_M PANDFVRGFVGAGATLMQLSLTRVRDIDLHEAVVAQAGGDPAEAVELAGS
MUL_0033|M.ulcerans_Agy99 PANDFVRGFVGAGATLMQLSLTRVRDIDLHEAVVAQAGGDPAEAVELAGS
TH_4520|M.thermoresistible__bu PADDFVKGFVGHGAALKQLTLTRVRDVDLHDAVTARAGGDPDEAIHAARA
Mflv_3109|M.gilvum_PYR-GCK PADDTVAGFVGSGASLQQLGLLRVKDVALEPRPSVHRGDSVEQARAVIGA
MLBr_01424|M.leprae_Br4923 PENVFVAGFIGS-PGMNLVTLSIVDSSVLLGDWPIRIPREIASAASEVII
* : * *:* : : . . : : .
Mb3784c|M.bovis_AF2122/97 RDG-WVLVVDGAGAPLGWIDADGRRRHRGGAALSDAMTVGGSVFRPNGNL
Rv3758c|M.tuberculosis_H37Rv RDG-WVLVVDGAGAPLGWIDADGRRRHRGGAALSDAMTVGGSVFRPNGNL
MAV_0318|M.avium_104 FDG-WAVVVDEAGAPLGWIDAEGLRRHRDGLALSDSMTGVGSLFRPRGNL
Mvan_5600|M.vanbaalenii_PYR-1 NPGDWVLVTRTDGTPYAWINAAGVELHRNGSSLYDSTIAGGSLFQPDGTL
MSMEG_2926|M.smegmatis_MC2_155 IDREHVIVLDQHDRPLRWLSLS--ELSDAHSLERVTRDENLEKVSTSSTL
MAB_1226|M.abscessus_ATCC_1997 IDREHVIVLDSHERPLRWSSLD--ELRAPNALQRVSQDEDLEKVSLASTL
MMAR_0034|M.marinum_M LDREHAIVLDRQNRPLRWLPLE--ELAKPQALAQVTRDDDLECVNLASTL
MUL_0033|M.ulcerans_Agy99 LDRAHAIVLDRQNRPLRWLPLE--ELAKPQALAQVTRDDDLECVNLASTL
TH_4520|M.thermoresistible__bu ADREHVIVLDAQNRPMRWMSLA--ELARPGALIRVSRDENLECVTLASTL
Mflv_3109|M.gilvum_PYR-GCK SDHDWAVVLDDRDRPVNWVRES--RLRHAGALADAV--EKLDVVSTYSTL
MLBr_01424|M.leprae_Br4923 GVRPEHFELGSLG-----VEVEIDMVEELGADAYLYGRIAGASKVTDQLV
. :
Mb3784c|M.bovis_AF2122/97 SQALDAALSSPSGVGVAVDGGGKVIGGILAADVLAEFQKGKKAGGGAKPC
Rv3758c|M.tuberculosis_H37Rv SQALDAALSSPSGVGVAVDGGGKVIGGILAADVLAEFQKGKKAGGGAKPC
MAV_0318|M.avium_104 SQALDAALSSPSGIGVAVDDDGRVIGGVLAADVLAAVESQRKSR------
Mvan_5600|M.vanbaalenii_PYR-1 RLALDAALSSPAGLGVAVDGAGQVIGGVRADDVLAALDKQRAARQAEG--
MSMEG_2926|M.smegmatis_MC2_155 NDALDVMLTSSHGVVVVTGRRNAYQGVIRVETIMDAMPDMQTSPTGPVL-
MAB_1226|M.abscessus_ATCC_1997 TDALEDMLTSSHGVVVVTGRRNTYQGVIRVETIMDAMAEMRAAAQ-----
MMAR_0034|M.marinum_M NDALDEMLTSSHGVVVVTGRRNTYQGVIRVETIMEAIADIRGAANSEEPD
MUL_0033|M.ulcerans_Agy99 NDALDEMLTSAHGVVVVTGRRNTYQGVIRVETIMEAIADIRGAANSEEPD
TH_4520|M.thermoresistible__bu TDALDDMLTSSHGVVVVTGRRNTYQGVIRLETIMDAIADARGTAGSQGAG
Mflv_3109|M.gilvum_PYR-GCK EDALEAILAEQHASAVVAGPNNRYEGVVTLDTLIDTITRLRAEAEASTGA
MLBr_01424|M.leprae_Br4923 VARVDGRNPPAKGSRVRLYTEPGNVHFFGVDGHRIS--------------
:: . . * .
Mb3784c|M.bovis_AF2122/97 TT--
Rv3758c|M.tuberculosis_H37Rv TT--
MAV_0318|M.avium_104 ----
Mvan_5600|M.vanbaalenii_PYR-1 ----
MSMEG_2926|M.smegmatis_MC2_155 ----
MAB_1226|M.abscessus_ATCC_1997 ----
MMAR_0034|M.marinum_M T---
MUL_0033|M.ulcerans_Agy99 T---
TH_4520|M.thermoresistible__bu AP--
Mflv_3109|M.gilvum_PYR-GCK GDPS
MLBr_01424|M.leprae_Br4923 ----