For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDPISVADPEHARALYEPLTQSVRRLVDLTIRSEADDDAVRRAHRLVDEAAALLGSRVAPGSFGIRSDAA GRPMPWGNVAIGVRNAIAPPLDVHRDSDGRATADLVLGAPYEGPPGQVHGGVCALVLDHVLGATAHHRHQ TAFTGTLTIRYVRPTWLGALRAEAWIDREEGSKTYAVGHLLGPDGEVTAQAEGVFIRPRSAR*
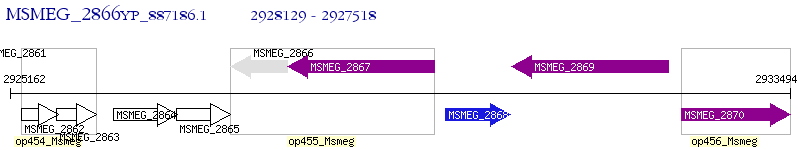
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2866 | - | - | 100% (203) | thioesterase family protein |
| M. smegmatis MC2 155 | MSMEG_3571 | - | 5e-57 | 58.50% (200) | thioesterase family protein |
| M. smegmatis MC2 155 | MSMEG_2929 | - | 2e-12 | 31.34% (201) | thioesterase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0363c | - | 2e-05 | 24.87% (193) | hypothetical protein Mb0363c |
| M. gilvum PYR-GCK | Mflv_3328 | - | 6e-53 | 53.73% (201) | thioesterase superfamily protein |
| M. tuberculosis H37Rv | Rv0356c | - | 2e-05 | 24.87% (193) | hypothetical protein Rv0356c |
| M. leprae Br4923 | MLBr_00279 | - | 1e-05 | 25.56% (180) | hypothetical protein MLBr_00279 |
| M. abscessus ATCC 19977 | MAB_4248c | - | 2e-08 | 26.97% (178) | hypothetical protein MAB_4248c |
| M. marinum M | MMAR_0247 | - | 5e-52 | 47.55% (204) | hypothetical protein MMAR_0247 |
| M. avium 104 | MAV_2246 | - | 3e-48 | 49.02% (204) | thioesterase family protein |
| M. thermoresistible (build 8) | TH_0396 | - | 1e-72 | 67.17% (198) | thioesterase family protein |
| M. ulcerans Agy99 | MUL_4866 | - | 6e-53 | 48.04% (204) | hypothetical protein MUL_4866 |
| M. vanbaalenii PYR-1 | Mvan_2864 | - | 1e-67 | 59.50% (200) | thioesterase superfamily protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2866|M.smegmatis_MC2_155 ----------------------------------------------MSDP
TH_0396|M.thermoresistible__bu ----------------------------------------------MPEQ
Mvan_2864|M.vanbaalenii_PYR-1 ----------------------------------------------MSRP
MMAR_0247|M.marinum_M ----------------------------------------------MSDE
MUL_4866|M.ulcerans_Agy99 ----------------------------------------------MSDE
Mflv_3328|M.gilvum_PYR-GCK -----------------------------------------MLDYTHVDG
MAV_2246|M.avium_104 -------------------------------------------MDAIFEV
Mb0363c|M.bovis_AF2122/97 ---------------------MT-----DASVHPDELDP----EYHHHGG
Rv0356c|M.tuberculosis_H37Rv ---------------------MT-----DASVHPDELDP----EYHHHGG
MLBr_00279|M.leprae_Br4923 ---------------------MTY----YSSLRPEDLPPE-RPKHEHYSG
MAB_4248c|M.abscessus_ATCC_199 MHWRRAGPGSNDREAGHTLAVMTADGDRLAALHEEMNRAIREIDGEHSGG
MSMEG_2866|M.smegmatis_MC2_155 ISVADPEHARALYEPLTQSVRRLVDLTIRSEADDDAVRRAHRLVDEAAAL
TH_0396|M.thermoresistible__bu VPVSRAEQDRARYEPLTSSVRRLIDATIRSEADEQAVRQAHALIDQAVAL
Mvan_2864|M.vanbaalenii_PYR-1 LRPEEVQRAEDLFGPLTQALRTLVDVAIRSDADDPDVTRAHELIEQATEL
MMAR_0247|M.marinum_M WPGTDFDRLESVYGPLTASIRRLIDVSIRTQADPATVTASMAKIDSAAEE
MUL_4866|M.ulcerans_Agy99 CPGTDFDRLESVYGPLTASIRRLIDVSIRTQADPATVTASMAKIDSAAEE
Mflv_3328|M.gilvum_PYR-GCK LSAADIARRRELYEPLTASVRDLIDATILTEADPDAVAAAKAEIESATAR
MAV_2246|M.avium_104 LDAAEAERVTALYAPLADAVRELVDATIRTEVDDAVVAEARSAIEAVTAS
Mb0363c|M.bovis_AF2122/97 FPEYGPASPGAGFGQFVATMRRLQDLAVAADPGDAVWDEAAERAAALVEL
Rv0356c|M.tuberculosis_H37Rv FPEYGPASPGAGFGQFVATMRRLQDLAVAADPGDAVWDEAAERAAALVEL
MLBr_00279|M.leprae_Br4923 FPEYELANPGVGFRRFVATMRRLQDLAVSADPSDEVWYAAADRAVALVEL
MAB_4248c|M.abscessus_ATCC_199 FPQYSPSSVGPGFGRFLTAMRRLQDLAVSADMDDPRWDEAADLTEKLVEL
: : ::* * * :: :: . : .
MSMEG_2866|M.smegmatis_MC2_155 LGSRVAPGSFGIRSDAAGRPMPWGNVAIGVRNAIAPPLDVHRDSDGRATA
TH_0396|M.thermoresistible__bu LSSRLMPGSFGLRHDTDGNPMVWGNVAIGLRNAIAPPLRVHHQG-GRAWA
Mvan_2864|M.vanbaalenii_PYR-1 LSTRLEPEPFPVRTTTTGRILTWGNVAIGMRNAVAPPLRLQRDDTGRVTT
MMAR_0247|M.marinum_M LSDALHPGGFGVQLTPEGETVAWGNVVVGVRNPVAPPLSIHHEPNGLVWS
MUL_4866|M.ulcerans_Agy99 LSDALHPGGFGVQLTPEGETVAWGNVVVGVRNPVAPPLSIHHEPDGLVWS
Mflv_3328|M.gilvum_PYR-GCK LRSAARDGSFGIQFGADGESMPWGNAVIGVRNPTAPPLVIHKEPDGFARS
MAV_2246|M.avium_104 LRRRTRP--VGVSYRVDGRPLPLGNAAIGVCNPIAPPIVVHHEGDGRCWS
Mb0363c|M.bovis_AF2122/97 LSPFEADEGKAPAGRTPG--------LPGMGSLLLPPWTVTRYGTDGVEM
Rv0356c|M.tuberculosis_H37Rv LSPFEADEGKAPAGRTPG--------LPGMGSLLLPPWTVTRYGTDGVEM
MLBr_00279|M.leprae_Br4923 LGPFATDEGKAPAGRVPD--------MPGMGSLLLPPWTLTRSGPDSVEM
MAB_4248c|M.abscessus_ATCC_199 MDPFEAPEGVGPANRVAD--------EPANGHLLLPPWFIDKAGPDGIEA
: . . ** : : .
MSMEG_2866|M.smegmatis_MC2_155 DLVLGAPYEGPPGQVHGGVCALVLDHVLGATAHH-RHQTAFTGTLTIRYV
TH_0396|M.thermoresistible__bu EVTLGAPYEGPPGQVHGGVCALLLDHVLGATAHR-PGAPAFTGTITLRYL
Mvan_2864|M.vanbaalenii_PYR-1 DLTLGVPYEGPAGHVHGGVCALLLDHVLSATAHR-RGEPAFTGTLNVRYV
MMAR_0247|M.marinum_M EFTLGAAYEGPPGHVHGGVCSMILDHVLGATAHL-PRRPAYTGTLTLRYR
MUL_4866|M.ulcerans_Agy99 EFTLGAAYEGPPGHVHGGVCSMILDHVLGATAHL-PRRPAYTGTLTLRYR
Mflv_3328|M.gilvum_PYR-GCK DFYLGAAFEGPPGHVHGGVSAKILDHVLGDAASK-PGVHRLTGTITVRYR
MAV_2246|M.avium_104 EFVLGSAYEGPPRLVHGGVSALVLDHMLGEAASEGLSKARFTGTITVKYL
Mb0363c|M.bovis_AF2122/97 RGSFSRFHVGGNSAVHGGVLPLLFDHMFGMISHAAGRPISRTAFLHVDYR
Rv0356c|M.tuberculosis_H37Rv RGSFSRFHVGGNSAVHGGVLPLLFDHMFGMISHAAGRPISRTAFLHVDYR
MLBr_00279|M.leprae_Br4923 TGYFTRFHVGFNHAVHGGVLPLVFDHLFGMISYTAGRSISRTAFLHVDYR
MAB_4248c|M.abscessus_ATCC_199 HGDFPRFYLGGNGAVHGGILPLLFDHLFGMTVIMAGRTISRTAFLHVNYR
: . * ****: . ::**::. *. : : *
MSMEG_2866|M.smegmatis_MC2_155 RPTWLGA-LRAEAWIDREEGSKTYAVGHLLGPDGEVTAQAEGVFIRPRSA
TH_0396|M.thermoresistible__bu RATPLGVPLRTEAWVDREEGSKTFALGQISDHDGVVTVRGEGVFIRPG--
Mvan_2864|M.vanbaalenii_PYR-1 APTRLGR-LRVQAWTEREEGAKTFAAGHITDADGAVTVQAEGVFIRPKVK
MMAR_0247|M.marinum_M RGTVLGRPLRAEGRVERIEGVKTFSVGHIADENG-ITVEAEGIFIQSKQI
MUL_4866|M.ulcerans_Agy99 RGTVLGRPLRAEGRVERIEGVKTFSVGHIADESG-ITVEAEGIFIQSKQI
Mflv_3328|M.gilvum_PYR-GCK RLTPLGR-LHSEARVTHTDGFKTYAVGHIADATG-ITVEAEAVFIEPRWA
MAV_2246|M.avium_104 RGTPLGP-LRCDAWIDRREGVKVFARGIISDAAG-VTVEADGVFIEPAWA
Mb0363c|M.bovis_AF2122/97 RITPIDVPLIVRGRVTNTEGRKAFVCAELFDSDETLLAEGNGLMVRLLPG
Rv0356c|M.tuberculosis_H37Rv RITPIDVPLIVRGRVTNTEGRKAFVCAELFDSDETLLAEGNGLMVRLLPG
MLBr_00279|M.leprae_Br4923 KITPIDEPLVMRGRVTRTEGRKAFVSAELVDGDEMLLAEGNGMMVRLLAG
MAB_4248c|M.abscessus_ATCC_199 QVVPVGVPLTASSRIDEVDRRKAFVSAELCDAHGVVLADANGLMVQLLPG
. :. * . . : *.: . : . : . .:.:::.
MSMEG_2866|M.smegmatis_MC2_155 R---
TH_0396|M.thermoresistible__bu ----
Mvan_2864|M.vanbaalenii_PYR-1 S---
MMAR_0247|M.marinum_M RA--
MUL_4866|M.ulcerans_Agy99 RA--
Mflv_3328|M.gilvum_PYR-GCK RG--
MAV_2246|M.avium_104 RETQ
Mb0363c|M.bovis_AF2122/97 QP--
Rv0356c|M.tuberculosis_H37Rv QP--
MLBr_00279|M.leprae_Br4923 QP--
MAB_4248c|M.abscessus_ATCC_199 QP--