For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRIAVVAGPDPGHAFPALALCLKFLAAGDQPTLLTGVEWLQTARDAGVDAVELDGLDPTDEDDDADAGAK IHRRAARMAVLNTPRLRDLAPDLIVSDVITACGGMAAELLGLPWVELNPHPLYLPSRGLPPVGSGLAPGV GLRGKLRDAIMRALTARSWKAGLAQRAQARAGIGLPAEDPGPLRRLIATLPALEVPRPDWPHEAVVVGPL HFEPTSAVLQIPAGEGPVVVVAPSTAHTGAQGMAELALECLRPGETLPPGARVVVSRLAGPDTEVPPWAA VGLGRQDELLTHADVVICGGGHGMVSKTLLAGVPMVVVPGGGDQWEIANRVARQGSAQLVRPLTGDSLTA AVGEVLGSPGYRRAAAAAGASVAEVADPVQVCHDALMRTG
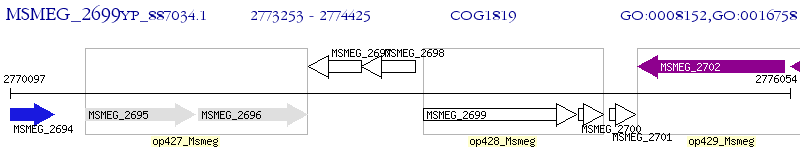
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2699 | - | - | 100% (390) | hypothetical protein MSMEG_2699 |
| M. smegmatis MC2 155 | MSMEG_2342 | - | 3e-28 | 31.30% (409) | putative glycosyltransferase |
| M. smegmatis MC2 155 | MSMEG_4670 | - | 7e-15 | 27.21% (419) | N-glycosylation |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2759c | - | 1e-173 | 76.41% (390) | alanine rich transferase |
| M. gilvum PYR-GCK | Mflv_3984 | - | 1e-178 | 78.24% (386) | UDP-glucuronosyl/UDP-glucosyltransferase |
| M. tuberculosis H37Rv | Rv2739c | - | 1e-173 | 76.41% (390) | alanine rich transferase |
| M. leprae Br4923 | MLBr_00985 | - | 1e-165 | 73.85% (390) | putative glycosyltransferase |
| M. abscessus ATCC 19977 | MAB_3062c | - | 1e-158 | 71.24% (386) | hypothetical protein MAB_3062c |
| M. marinum M | MMAR_1974 | - | 1e-175 | 76.41% (390) | glycosyl transferase |
| M. avium 104 | MAV_3631 | - | 1e-179 | 78.76% (386) | hypothetical protein MAV_3631 |
| M. thermoresistible (build 8) | TH_2807 | - | 6e-33 | 79.76% (84) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3383 | - | 1e-174 | 76.41% (390) | glycosyl transferase |
| M. vanbaalenii PYR-1 | Mvan_2401 | - | 1e-179 | 78.76% (386) | UDP-glucuronosyl/UDP-glucosyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3984|M.gilvum_PYR-GCK ----MRVAVVAGPDPGHAFPALELCRRFRAAGDAPTLLTGTRWLDVARGE
Mvan_2401|M.vanbaalenii_PYR-1 ----MRVAVVAGPDPGHAFPAIALCLRFLAAGDHPTLLTGTKWLDTARRE
MMAR_1974|M.marinum_M ----MRVAVVAGPDPGHSFPAIALCRRFLAAGDIPTLLTGVEWLDTARAA
MUL_3383|M.ulcerans_Agy99 ----MRVAVVAGPDPGHSFPAIALCRRFLAAGDIPTLLTGVEWLDTARAA
Mb2759c|M.bovis_AF2122/97 ----MRVAVVAGPDPGHSFPAIALCQRFRAAADTPTLFTGVEWLEAARAA
Rv2739c|M.tuberculosis_H37Rv ----MRVAVVAGPDPGHSFPAIALCQRFRAAADTPTLFTGVEWLEAARAA
MLBr_00985|M.leprae_Br4923 MVHYMRVAVVAGPDPGHSFPAIALCQRFAEAGDTPTLFTGVECVDIARAA
MAV_3631|M.avium_104 ----MRVAVVAGPDPGHSFPAIALCRRFADAGDTPTLFTGAEWLDTARGA
TH_2807|M.thermoresistible__bu --------------------------------------------------
MSMEG_2699|M.smegmatis_MC2_155 ----MRIAVVAGPDPGHAFPALALCLKFLAAGDQPTLLTGVEWLQTARDA
MAB_3062c|M.abscessus_ATCC_199 ----MRVAVVAGPDPGHAFPALALCLKFAAAGDDPVLLTGTRWLDQARAA
Mflv_3984|M.gilvum_PYR-GCK GVDAVELDGLDPTAFDDDTDAGEKIHRRAARMAVLNRARIGELSPDLVVS
Mvan_2401|M.vanbaalenii_PYR-1 GVDAVELLGLDPTADDDDTDAGAKIHHRAARMAVLNKDPIAALAPDLVIS
MMAR_1974|M.marinum_M GVDAIELAGLVATDED--LDAGARIHRRAAQMALLNAPALSDLAPDLVVS
MUL_3383|M.ulcerans_Agy99 GVDAIELAGLVATDED--LDAGARIHRRAAQMALLNAPALSDLAPDLVVS
Mb2759c|M.bovis_AF2122/97 GIDAVELDGLAATDRD--LDAGARIHRRAAQMAVLNVPRLRALEPELVVS
Rv2739c|M.tuberculosis_H37Rv GIDAVELDGLAATDRD--LDAGARIHRRAAQMAVLNVPRLRALEPELVVS
MLBr_00985|M.leprae_Br4923 GVETALLDGLVAVDDD--VDAGARIHRRAAQMAVLNVPVLRDLAPDLIVS
MAV_3631|M.avium_104 GVDAVELDGLAATDED--VDAGARIHRRAARMAVLNVPALRDLAPDLVVS
TH_2807|M.thermoresistible__bu --------------------------------------------------
MSMEG_2699|M.smegmatis_MC2_155 GVDAVELDGLDPTDEDDDADAGAKIHRRAARMAVLNTPRLRDLAPDLIVS
MAB_3062c|M.abscessus_ATCC_199 GVESVELLGLQPRVDDDDADSGAKIHQRAAHMALANLDQLRALAPDLIVS
Mflv_3984|M.gilvum_PYR-GCK DSITVCGGLAADLLGVPWVELNTHPLYRPSRGLPPVGSGLAPGVGLRGRA
Mvan_2401|M.vanbaalenii_PYR-1 DSITTCGGLAADLLGLPWAELNTHPLYHPSKGLPPIGSGLAPGTGLRGRL
MMAR_1974|M.marinum_M DVITACGGMAAERLGIPWIELNPHPLYRPSKGLPPIGSGLAPGTGIRGRL
MUL_3383|M.ulcerans_Agy99 DVITACGGMAAERLGIPWIELNPHPLYRPSKGLPPIGSGLAPGTGIRGRL
Mb2759c|M.bovis_AF2122/97 DVITACGGMAAELLGIPWVELNPHPLYLPSKGLPPIGSGLAAGTGIRGRL
Rv2739c|M.tuberculosis_H37Rv DVITACGGMAAELLGIPWVELNPHPLYLPSKGLPPIGSGLAAGTGIRGRL
MLBr_00985|M.leprae_Br4923 DVITAAGGMAAELLGIPWIELNPHPLYLPSKGLPPIGSGLASGVGIHGRM
MAV_3631|M.avium_104 DVITAGGGMAAELLGIPWIELSPHPLYLPSKGLPPIGSGLATGTGLRGRL
TH_2807|M.thermoresistible__bu --------------------------------------------------
MSMEG_2699|M.smegmatis_MC2_155 DVITACGGMAAELLGLPWVELNPHPLYLPSRGLPPVGSGLAPGVGLRGKL
MAB_3062c|M.abscessus_ATCC_199 DVITACGGMAGELLGVPWVELNPHPLYLPSKGLPPLGSGLAPGVGLRGRL
Mflv_3984|M.gilvum_PYR-GCK RDAVLRALTARSWRAGLRQRAAARVEIGLPGDDPGPVRRLIATLPALEVP
Mvan_2401|M.vanbaalenii_PYR-1 RDAVLRALSARSWRAGLRQRSEARVGIGLPAADPGPVCRLIATLPALEVP
MMAR_1974|M.marinum_M RDTTMRALTARSWRAGLRQRAAVRAEIGLAEHDPGPLRRLIATLPALEVP
MUL_3383|M.ulcerans_Agy99 RDTTMRALTARSWRAGLRQRAAVRAEIGLAEHDPGPLRRLIATLPALEVP
Mb2759c|M.bovis_AF2122/97 RDATMRALTGRSWRAGLRQRAAVRVEIGLPARDPGPLRRLIATLPALEVP
Rv2739c|M.tuberculosis_H37Rv RDATMRALTGRSWRAGLRQRAAVRVEIGLPARDPGPLRRLIATLPALEVP
MLBr_00985|M.leprae_Br4923 RDATMRALTARSWRAGLRERAAVRVQIGLPACDSGPLRRLIATLPALEVS
MAV_3631|M.avium_104 RDATMRALTARSWRAGLRQRAAARAEIGLPARDPGPLRRLIATLPALEVP
TH_2807|M.thermoresistible__bu --------------------------------------------------
MSMEG_2699|M.smegmatis_MC2_155 RDAIMRALTARSWKAGLAQRAQARAGIGLPAEDPGPLRRLIATLPALEVP
MAB_3062c|M.abscessus_ATCC_199 RDGLMRRLTQKSIDQGLRQRSQARVEIGLPARDPGPMRRLIATLPALEVP
Mflv_3984|M.gilvum_PYR-GCK RPDWPAEAVVVGPLHFEPTTELLAVPAGGGPLVVVAPSTATTGAGGMAEL
Mvan_2401|M.vanbaalenii_PYR-1 RPDWPADAVVVGPLHFEPTTHVLRLPAGDGPVVVVAPSTATTGAVGLAEL
MMAR_1974|M.marinum_M RPDWPAEAIVVGPLHFEPTDQLLKVPAGSGPVLVIAPSTALSGTAGLAEV
MUL_3383|M.ulcerans_Agy99 RPDWPAEAIVVGPLHFEPTDQLLKVPAGSGPVLVIAPSTALSGTAGLAEV
Mb2759c|M.bovis_AF2122/97 RPDWPAEAVVVGPLHFEPTDRVLAIPAGTGPVVVVAPSTALTGTAGLTEV
Rv2739c|M.tuberculosis_H37Rv RPDWPAEAVVVGPLHFEPTDRVLAIPAGTGPVVVVAPSTALTGTAGLTEV
MLBr_00985|M.leprae_Br4923 RPDWPAEAVVVGPLHFEPTDRVLQIPSGSGPVIVVAPSTALTGISGMAQT
MAV_3631|M.avium_104 RPDWPDEAVLVGPLHFEPTDRVLDIPAGSGPVVVVAPSTAQTGARGLAEV
TH_2807|M.thermoresistible__bu --------------------------------------------------
MSMEG_2699|M.smegmatis_MC2_155 RPDWPHEAVVVGPLHFEPTSAVLQIPAGEGPVVVVAPSTAHTGAQGMAEL
MAB_3062c|M.abscessus_ATCC_199 RPDWPAEAVVVGPLHWEPTDVVLQPPSGSGPVVVIAPSTAFTGAEGMTEL
Mflv_3984|M.gilvum_PYR-GCK ALNTLVPGDTLPPGARVAVSQLDGPDSAVPPWAVVGLGRQDELLARADLL
Mvan_2401|M.vanbaalenii_PYR-1 ALATLVPGEVLPEGARVVVSRLEGPDAEVPPWAVVGLGRQDELLAKADLL
MMAR_1974|M.marinum_M ALESLIPGDTLPSGSRLVVSRLGGDDLVTPSWAAVGLGSQAELLTHADVV
MUL_3383|M.ulcerans_Agy99 ALESLIPGGTLSSGSRLVVSRLGGDDLVTPSWAAVGLGSQAELLTHADVV
Mb2759c|M.bovis_AF2122/97 ALQSLTPGETVPSGSRLVVSRLSGADLTVPPWAVAGLGSQAELLTRADLV
Rv2739c|M.tuberculosis_H37Rv ALQSLTPGETVPSGSRLVVSRLSGADLTVPPWAVAGLGSQAELLTRADLV
MLBr_00985|M.leprae_Br4923 ALDALVPGDTLPSGSRVVVSRLNGPDLTVPSWAVAGLVHQTKLLAHADVV
MAV_3631|M.avium_104 ALSCLTPGETLPAGARLVVSRLAGPELAAPPWAVVGLGSQAELLRHADVV
TH_2807|M.thermoresistible__bu --------------------------------------------------
MSMEG_2699|M.smegmatis_MC2_155 ALECLRPGETLPPGARVVVSRLAGPDTEVPPWAAVGLGRQDELLTHADVV
MAB_3062c|M.abscessus_ATCC_199 AVQALES-----AGARVVISALEPPSSGLPSWAIAGLARQDLLLSGADLA
Mflv_3984|M.gilvum_PYR-GCK ICGGGHGTVAKSLLAGVPMVVVPGGGDQWEIANRLVRQGSAQLVRPLSAE
Mvan_2401|M.vanbaalenii_PYR-1 ICGGGHGTVAKSLLAGVPMVVVPGGGDQWEIANRLVRQGSAQLVRPLTGS
MMAR_1974|M.marinum_M ICGGGHGMVSKTLLAGVPLVVVPGGGDQWEIANRVVRQGSARLIRPLGAE
MUL_3383|M.ulcerans_Agy99 ICGGGHGMVSKTLLAGVPLVVVPGGGDQWEIANRVVRQGSARLIRPLGAD
Mb2759c|M.bovis_AF2122/97 ICGGGHGMVAKTLLAGVPMVVVPGGGDQWEIANRVVRQGSAVLIRPLTAD
Rv2739c|M.tuberculosis_H37Rv ICGGGHGMVAKTLLAGVPMVVVPGGGDQWEIANRVVRQGSAVLIRPLTAD
MLBr_00985|M.leprae_Br4923 ICGGGHGMVAKTLLAGVPLVVVPGGGDQWEMANRVVRQGSAQLIRPLTAA
MAV_3631|M.avium_104 VCGGGHGMVAKTLLAGVPLVAVPGGGDQWEIANRVVRQGSARLIRPLSAD
TH_2807|M.thermoresistible__bu -------MVSKTLLAGVPMVVVPGGGDQWEIANRVVRQGSGRLVRPLTGD
MSMEG_2699|M.smegmatis_MC2_155 ICGGGHGMVSKTLLAGVPMVVVPGGGDQWEIANRVARQGSAQLVRPLTGD
MAB_3062c|M.abscessus_ATCC_199 VCGGGHGFLSKALLAGVPTVVVPGGGDQWELANRVVRQGSAELVRPLSRD
::*:****** *.*********:***:.****. *:***
Mflv_3984|M.gilvum_PYR-GCK SLAAAVREVLGSPSYREAAVRAGAGVADVADPVRVCHDALGAPRKLGPCG
Mvan_2401|M.vanbaalenii_PYR-1 TLTAAVQEVLGSPGYREAARRAGAGVADVADPVRVCHAALGAPA------
MMAR_1974|M.marinum_M ALAAAVNEVLSSPGYREAAQRAAASIADVADPVQVCHEALALAG------
MUL_3383|M.ulcerans_Agy99 ALAAAVNEVLSSPGYREAAQRAAASIADVADPVQVCHEALALAG------
Mb2759c|M.bovis_AF2122/97 ALVAAVNEVLSSPRFREAARRAAASVAGAADPVRVCHDALALAG------
Rv2739c|M.tuberculosis_H37Rv ALVAAVNEVLSSPRFREAARRAAASVAGAADPVRVCHDALALAG------
MLBr_00985|M.leprae_Br4923 ALLAAVNKVLSSPDYREAAQRAAISIAGVADPVQVCHEALGLAG------
MAV_3631|M.avium_104 ALVAAVNEVLSSPGYRAAAQRAAAGIADVADPVRVCREALAG--------
TH_2807|M.thermoresistible__bu ALVAAVGTVLSDPGYRAAARRAGASVADVEDPVRVCQEALIGSA------
MSMEG_2699|M.smegmatis_MC2_155 SLTAAVGEVLGSPGYRRAAAAAGASVAEVADPVQVCHDALMRTG------
MAB_3062c|M.abscessus_ATCC_199 SLASAIGRVLGEPRYREAARRAAAGAIAVEDPVRVCQDVLAALR------
:* :*: **..* :* ** *. . . ***:**: .*