For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTDQQASLVASKPVGEPQKKQSRAGRNLPAAIGVGVGLGGGVIAILLFAPLVWVGVVAAAMAVATHEVVR RLRTGGFSIPVVPLLIGGQAMVWLTLPFGPAGALGGFGATVVLCLIWRLLSGGLTAAPVNYLRDVSVTVF LAAWIPLFGAFGALLIYPDDGAGRVFCLMLGVVASDVGGYTAGVLFGKHPMVPAISPKKSWEGLAGSLVL GIVVSVLAVTFLLDKPAWVGVPLGIMLVITGTLGDLVESQVKRDLGIKDMGTLLPGHGGLMDRIDSVLPS AVATWIVLTLLA
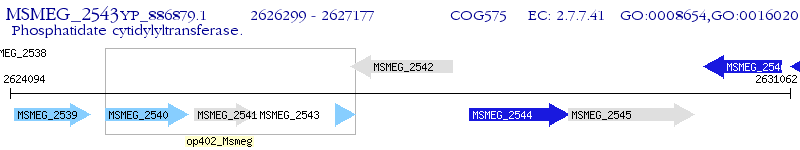
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2543 | cdsA | - | 100% (292) | phosphatidate cytidylyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2905c | cdsA | 1e-107 | 65.14% (284) | integral membrane phosphatidate cytidylyltransferase CdsA |
| M. gilvum PYR-GCK | Mflv_4122 | - | 1e-125 | 73.29% (292) | phosphatidate cytidylyltransferase |
| M. tuberculosis H37Rv | Rv2881c | cdsA | 1e-107 | 65.14% (284) | integral membrane phosphatidate cytidylyltransferase CdsA |
| M. leprae Br4923 | MLBr_01589 | - | 1e-105 | 66.20% (284) | putative phosphatidate cytidylyltransferase |
| M. abscessus ATCC 19977 | MAB_3186c | - | 1e-113 | 67.71% (288) | phosphatidate cytidylyltransferase (CdsA) |
| M. marinum M | MMAR_1828 | cdsA | 1e-106 | 62.67% (300) | integral membrane phosphatidate cytidylyltransferase CdsA |
| M. avium 104 | MAV_3731 | cdsA | 1e-107 | 69.15% (282) | phosphatidate cytidylyltransferase |
| M. thermoresistible (build 8) | TH_0273 | cdsA | 1e-115 | 68.47% (295) | PROBABLE INTEGRAL MEMBRANE PHOSPHATIDATE |
| M. ulcerans Agy99 | MUL_2076 | cdsA | 1e-107 | 62.67% (300) | integral membrane phosphatidate cytidylyltransferase CdsA |
| M. vanbaalenii PYR-1 | Mvan_2222 | - | 1e-139 | 81.16% (292) | phosphatidate cytidylyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2543|M.smegmatis_MC2_155 --MTDQQASLVASKPVG---------EPQKKQSRAGRNLPAAIGVGVGLG
Mvan_2222|M.vanbaalenii_PYR-1 --MTDQHSTPVISKPVN---------EPPKKASRAGRNLPAAIAVGVALG
Mflv_4122|M.gilvum_PYR-GCK --MTDQHSTPVGPQPVD---------EPPKKSSRAGRNLPAAIAVGVLLG
TH_0273|M.thermoresistible__bu -------------VPAD---------ET-QKVGRAGRNLPAAIAVGVLLG
MAB_3186c|M.abscessus_ATCC_199 --MGDTDAAPAPGSSPA---------PPAGAKSRAGRNLPAAIGVGVALA
Mb2905c|M.bovis_AF2122/97 MTTNDAGTGNPAEQPARGAKQ-----QPATETSRAGRDLRAAIVVGLSIG
Rv2881c|M.tuberculosis_H37Rv MTTNDAGTGNPAEQPARGAKQ-----QPATETSRAGRDLRAAIVVGLSIG
MMAR_1828|M.marinum_M MANTDATTGTPADEPVQ---------EPPKKASRAGRDLPAAIAVGLSIG
MUL_2076|M.ulcerans_Agy99 MANTDATTGTPADEPVQ---------EPPKKASRAGRDLPAAIAVGLSIG
MLBr_01589|M.leprae_Br4923 MASTDPGTGTPLDESVPGIKR--AMRQSTKNTPRAGRNLPAAIAVGLSIG
MAV_3731|M.avium_104 MATSDPGAGDEPEHAVENTTEGAAGQRAKKKTSRAGRDLRAAIAVGAGIG
. . ****:* *** ** :.
MSMEG_2543|M.smegmatis_MC2_155 GGVIAILLFAPLVWVGVVAAAMAVATHEVVRRLRTGGFSIPVVPLLIGGQ
Mvan_2222|M.vanbaalenii_PYR-1 AMLIVVLLFAPKVWVGVVSVAIAIATHEVVRRLREGGYSIPVVPLLVGGQ
Mflv_4122|M.gilvum_PYR-GCK GGLIAILIFAPYLWLALLSAAVAMATYEVSSRLGEAGYRIPFVPLLVGGQ
TH_0273|M.thermoresistible__bu GGLIAVLVWAPYVWLGVLAVAIAIATREVTARLREAGYQIPLVPLLLGGQ
MAB_3186c|M.abscessus_ATCC_199 ALIIFSLLYQKEIWVGIISVFLGIAMWEVTKRLRQADISVPFIPLFFGGQ
Mb2905c|M.bovis_AF2122/97 LVLIAVLVFVPRVWVAIVAVATLVATHEVVRRLREAGYLIPVIPLLIGGQ
Rv2881c|M.tuberculosis_H37Rv LVLIAVLVFVPRVWVAIVAVATLVATHEVVRRLREAGYLIPVIPLLIGGQ
MMAR_1828|M.marinum_M LFLVATLVFAPRGWVAICAFAMVVATHEVVRRLREAGYLIPVVPLLIGGQ
MUL_2076|M.ulcerans_Agy99 LFLVATLVFAPRGWVAICAFAMVVATHEVVRRLREAGYLIPVVPLLIGGQ
MLBr_01589|M.leprae_Br4923 GVLVATLVFAPRIWVVLCAGAIFVASHEVVRRLREAGYVIPAIPLLIGGQ
MAV_3731|M.avium_104 AVLIVTLVFAPRFWVPIVAMAILVASHEVVRRLREAGYVIPVIPLLAGGQ
:: *:: *: : : :* ** ** .. :* :**: ***
MSMEG_2543|M.smegmatis_MC2_155 AMVWLTLPFGPAGALGGFGATVVLCLIWRLLSGG-------------LT-
Mvan_2222|M.vanbaalenii_PYR-1 AMIWLTWPFGAAGALGAFGGTVVVCMIWRLVSGG-------------LS-
Mflv_4122|M.gilvum_PYR-GCK ATLWLTWPFGPSGALGGFAATVVVCMIWRLVGHG-------------LN-
TH_0273|M.thermoresistible__bu ATLWLTWPFGAAGALGGFAGMVVTCMIWRLIAQGRAAAPVRTSADPDLSG
MAB_3186c|M.abscessus_ATCC_199 AMIWLTWPWGTAGSVGAFAATVVVCMIWRLLSQG-------------LG-
Mb2905c|M.bovis_AF2122/97 AAVWLTWPFGAVGALAGFGGMVVVCMIWRLFMQDSVTRPTTG-------G
Rv2881c|M.tuberculosis_H37Rv AAVWLTWPFGAVGALAGFGGMVVVCMIWRLFMQDSVTRPTTG-------G
MMAR_1828|M.marinum_M LTVWLSWPYGVRGVLAGFGAMIVVCMFWRLFMQDNRPHDGAGS-----DG
MUL_2076|M.ulcerans_Agy99 LTVWLSWPYGVRGVLAGFGAMIVVCMFWRLFMQDNLPHDGAGS-----DG
MLBr_01589|M.leprae_Br4923 FTVWLTWPYRTVGALAGFGATVVVCMIWRLVMHDNSKQHESREA----LA
MAV_3731|M.avium_104 LTVWLTWPFHAAGALAGFGVTVVACLFWRLFMQDNRKRPEP-------FA
:**: *: * :..*. :* *::***. .
MSMEG_2543|M.smegmatis_MC2_155 -AAPVNYLRDVSVTVFLAAWIPLFGAFGALLIYPDDGAGRVFCLMLGVVA
Mvan_2222|M.vanbaalenii_PYR-1 -DAPQNYLRDMSTTVFLATWVPLFASFGVLLIYPDDGAGRVFCLMLGVVA
Mflv_4122|M.gilvum_PYR-GCK -HAPQNYTRDIGATVFLATWVPLFASFGALLIYPDDGANRVFCLMLGVVF
TH_0273|M.thermoresistible__bu QPGPTNYLRDIAATIFLATWVPLFAGFGALLIYPDDGPARVFSFMLAVVC
MAB_3186c|M.abscessus_ATCC_199 -SQPVNYLRDVAITVFVAAWIPLFASFGALLVYPDNGAEQVFCLMLGVTF
Mb2905c|M.bovis_AF2122/97 APSPGNYLSDVSATVFLAVWVPLFCSFGAMLVYPENGSGWVFCMMIAVIA
Rv2881c|M.tuberculosis_H37Rv APSPGNYLSDVSATVFLAVWVPLFCSFGAMLVYPENGSGWVFCMMIAVIA
MMAR_1828|M.marinum_M VPASGTYLRDTSATVFLAVWVVLFGSFAAMLVYPPNGTGWVFCVMIAIIA
MUL_2076|M.ulcerans_Agy99 VPASGTYLRDTSTTVFLAVWVVLFGSFAAMLVYPPNGTGWVFCVMIAIIA
MLBr_01589|M.leprae_Br4923 GPPVSNYLRDASATVFLAAWVPLFASFAALLVYPKDGAGRVFCLMIAVVA
MAV_3731|M.avium_104 GSPSANYLRDASATVFLACWVPLFASFAALLVYPADGAGRVFCLMITVVA
.* * . *:*:* *: ** .*..:*:** :*. **..*: :
MSMEG_2543|M.smegmatis_MC2_155 SDVGGYTAGVLFGKHPMVPAISPKKSWEGLAGSLVLGIVVSVLAVTFLLD
Mvan_2222|M.vanbaalenii_PYR-1 SDVGGYTAGVLFGKHPMAPAISPKKSWEGLAGSLVLGVVVSSLAVQFLTD
Mflv_4122|M.gilvum_PYR-GCK SDIGGYVAGVLFGRHTMAPAISPKKSWEGLAGSLLFGVTASVLAVVFLLD
TH_0273|M.thermoresistible__bu SDIGGYTAGVLFGRHPMVPAISPKKSWEGFAGSMVFGTTAAVLAVVFLMD
MAB_3186c|M.abscessus_ATCC_199 SDIGGYAAGVLFGKHPMAPAISPKKSWEGLAGSLVVGTTASTLAVTFLLG
Mb2905c|M.bovis_AF2122/97 SDVGGYAVGVLFGKHPMVPTISPKKSWEGFAGSLVCGITATIITATFLVG
Rv2881c|M.tuberculosis_H37Rv SDVGGYAVGVLFGKHPMVPTISPKKSWEGFAGSLVCGITATIITATFLVG
MMAR_1828|M.marinum_M SDVGGYTVGVLFGKHPMVPAISPKKSWEGFVGSLFCGIVATTITATFLAH
MUL_2076|M.ulcerans_Agy99 SDVGGYTVGVLFGKHPMVPTISPKKSWEGFAGSLFCGIVATTITATFLAH
MLBr_01589|M.leprae_Br4923 SDVGGYTVGVLFGKHPLVPRISPNKSWEGFAGSLVCGTTATILTATFLAG
MAV_3731|M.avium_104 SDVGGYAVGVLFGKHPMVPAISPKKSWEGLAGSLVLGITAATLAATFLAG
**:***..*****:*.:.* ***:*****:.**:. * ..: ::. **
MSMEG_2543|M.smegmatis_MC2_155 KPAWVGVPLGIMLVITGTLGDLVESQVKRDLGIKDMGTLLPGHGGLMDRI
Mvan_2222|M.vanbaalenii_PYR-1 NPWWIGIPLGLMLVITGTLGDLVESQVKRDLGIKDMGTLLPGHGGLMDRL
Mflv_4122|M.gilvum_PYR-GCK KPWWAGVALGLMLVITGTLGDLVESQFKRDLGIKDMSNLLPGHGGMMDRI
TH_0273|M.thermoresistible__bu KPWWAGAALGVLLVITATLGDLVESQFKRDLGIKDMGSLLPGHGGLMDRI
MAB_3186c|M.abscessus_ATCC_199 HHPFVGIALGVMLVITGTLGDLVESQVKRDLGIKDMGTLLPGHGGIMDRL
Mb2905c|M.bovis_AF2122/97 KTPWIGALLGVLFVLTTALGDLVESQVKRDLGIKDMGRLLPGHGGLMDRL
Rv2881c|M.tuberculosis_H37Rv KTPWIGALLGVLFVLTTALGDLVESQVKRDLGIKDMGRLLPGHGGLMDRL
MMAR_1828|M.marinum_M EPPWVGALLGVLFVLTTTLGDLVESQVKRDLGIKDMGRLLPGHGGLMDRL
MUL_2076|M.ulcerans_Agy99 EPPWVGALLGVLFVLTTTLGDLVESQVKRDLGIKDMGRLLPGHGGLMDRL
MLBr_01589|M.leprae_Br4923 KTPWVGALLSFVLVLTCTLGDLVESQVKRDLGIKDMGRLLPGHGGLMDRL
MAV_3731|M.avium_104 KAPWVGALLGVVLVFTCTLGDLVESQVKRDLGIKDMGRLLPGHGGLMDRL
. : * *..::*:* :********.*********. *******:***:
MSMEG_2543|M.smegmatis_MC2_155 DSVLPSAVATWIVLTLLA
Mvan_2222|M.vanbaalenii_PYR-1 DSILPSAVATWIVLSLLA
Mflv_4122|M.gilvum_PYR-GCK DAALPSAVATWIVLTLLA
TH_0273|M.thermoresistible__bu DGMLPSAVATWIVLSLLA
MAB_3186c|M.abscessus_ATCC_199 DSLLPSAVATWIVLTLFI
Mb2905c|M.bovis_AF2122/97 DGILPSAVAAWIVLTLLP
Rv2881c|M.tuberculosis_H37Rv DGILPSAVAAWIVLTLLP
MMAR_1828|M.marinum_M DGVLPSAVAAWIVMSLLT
MUL_2076|M.ulcerans_Agy99 DGVLPSAVAAWIVMSLLT
MLBr_01589|M.leprae_Br4923 DGVLPSAVVAWTMLTLLP
MAV_3731|M.avium_104 DGVLPSAVAAWTVLTLVP
*. *****.:* :::*.