For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PDTPDTLDRMPARTASAVTFLYALGYPIGNAAVAAMSPMALLVFRFSLAAAILGAWARFAGVAWPRGGKL AHVAVAGLLMQGVQFCFLYLAIERGAPAVLCAVVIAMNPVTTAVLSAAFLRDRLGLRRILALVLGVAAVL AACASRLAAESGVDPVLLLLLAALLGLSAGGVYQQRFCSDVDFRAATAVQNAVALLPAAVFAALTPFAVH NAKHAVAAVAGVVLLNAALGVSMYVRAISRHGAPAVTMLFCVIPAVAGVLSWLMLGERPDVGIAVGLVVG AAACWLNSSGSRQQRQHDPGRDG*
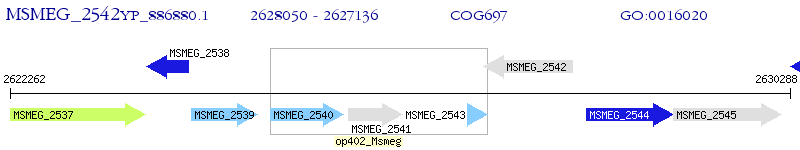
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2542 | - | - | 100% (304) | hypothetical protein MSMEG_2542 |
| M. smegmatis MC2 155 | MSMEG_2188 | - | 2e-10 | 25.86% (263) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_0513 | - | 4e-06 | 28.23% (209) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0187 | - | 5e-05 | 31.11% (180) | amino acid permease-associated region |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3180 | - | 9e-99 | 63.99% (286) | hypothetical protein MAB_3180 |
| M. marinum M | MMAR_4903 | - | 4e-08 | 25.33% (300) | hypothetical protein MMAR_4903 |
| M. avium 104 | MAV_0095 | - | 1e-107 | 66.55% (290) | hypothetical protein MAV_0095 |
| M. thermoresistible (build 8) | TH_0580 | rarD | 6e-06 | 25.28% (269) | RarD protein |
| M. ulcerans Agy99 | MUL_0503 | - | 4e-08 | 25.70% (284) | hypothetical protein MUL_0503 |
| M. vanbaalenii PYR-1 | Mvan_5539 | - | 3e-07 | 24.81% (262) | hypothetical protein Mvan_5539 |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_3180|M.abscessus_ATCC_1997 --------------------MQKTPVPMTKSSAVALTFLYALGYPIGALS
MAV_0095|M.avium_104 ---------------------------MPLTTASAVTFFYALGYPIGTLA
MSMEG_2542|M.smegmatis_MC2_155 -----------------MPDTPDTLDRMPARTASAVTFLYALGYPIGNAA
MMAR_4903|M.marinum_M ---------------------------MPALAMGVTVTLWAFAFVGVRSA
MUL_0503|M.ulcerans_Agy99 --------------MASSTTSSQRTVNMPALAMGVTVTLWAFAFVGVRSA
Mvan_5539|M.vanbaalenii_PYR-1 ------------------------MSLRGWLLFTAMGVIWGIPYLLIKVA
Mflv_0187|M.gilvum_PYR-GCK MRRRLGTFDTVTLGLGSMIGAGIFVALAPAAAAAGTGLLVGLAVAALIAV
TH_0580|M.thermoresistible__bu ---------------------MVNTRRQGLLCGVGAYLWWGLCPGYFPLL
.:
MAB_3180|M.abscessus_ATCC_1997 VSAMTPMLVLVAR------FALAGAILATWALIARALWPTGRTLCHVIVA
MAV_0095|M.avium_104 VVAMTPMAALVLR------FGLAAMVLAVWTAIAKAGWPRGARLGHVIVA
MSMEG_2542|M.smegmatis_MC2_155 VAAMSPMALLVFR------FSLAAAILGAWARFAGVAWPRGGKLAHVAVA
MMAR_4903|M.marinum_M GRVFSPGVLALSR------LFIAVMVMAVIATIAATYTAYAGRATTRRLR
MUL_0503|M.ulcerans_Agy99 GRVFSPGVLALSR------LFIAVMVMAVIATIAATYTAYAGRATTRRLR
Mvan_5539|M.vanbaalenii_PYR-1 VEGLSVPVLVFAR------TAVGALVLIPLTLRRGAWAPVLAHWRPVVAF
Mflv_0187|M.gilvum_PYR-GCK CNAMSSARLAARYPESGGTYVYGRERLGPFWGHTAGWSFIVGKTASCAAM
TH_0580|M.thermoresistible__bu APAGSVEVLAHRIVWS----VVFLLVVLTVARRLGDLVRLSARTWLYLAA
: :
MAB_3180|M.abscessus_ATCC_1997 GLLMQAVQFCALYEALEHGAPAVLG--------------AVVISMNPVVT
MAV_0095|M.avium_104 GLLIQGVQFCCLYEAVQLGAPAVLC--------------AVMIAMNPVAT
MSMEG_2542|M.smegmatis_MC2_155 GLLMQGVQFCFLYLAIERGAPAVLC--------------AVVIAMNPVTT
MMAR_4903|M.marinum_M PPSAAALTLVIAYGVAWFGAYSIVISWAEQ--HIDAGTTALLVNLAPILV
MUL_0503|M.ulcerans_Agy99 PPSAAALTLVIAYGVAWFGAYSIVISWAEQ--HIDAGTTALLVNLAPILV
Mvan_5539|M.vanbaalenii_PYR-1 AFFEIIAAWLLLSDAERHITSSLTG---------------LLIAAAPIVA
Mflv_0187|M.gilvum_PYR-GCK ALTVGYYVWPAYANAVAVGSVVALTAVSYAGVQRSATLTRVIVALVLGVL
TH_0580|M.thermoresistible__bu ASALISLNWGTYIYAVMNDHVVDAA---------------LGYFINPLVS
. :
MAB_3180|M.abscessus_ATCC_1997 AVLAALFLDER-LNRWRLVA-----------------LALGVVAVLAACA
MAV_0095|M.avium_104 AILAATFLREP-LGVQRTVA-----------------LVLGVLAVLAACS
MSMEG_2542|M.smegmatis_MC2_155 AVLSAAFLRDR-LGLRRILA-----------------LVLGVAAVLAACA
MMAR_4903|M.marinum_M AVFAGFFLGEG-YSARLLTG-----------------IAIAFAGVVLISA
MUL_0503|M.ulcerans_Agy99 AVFAGFFLGEG-YSARLLTG-----------------IAIAFAGVVLISA
Mvan_5539|M.vanbaalenii_PYR-1 ALLDRLTGGDQPLTVKRLAG-----------------LGTGLAGVAVLAG
Mflv_0187|M.gilvum_PYR-GCK AMVVVVVFGSGDVDASRLALGPDVDAYGVLQAAGLLFFAFAGYARIATLG
TH_0580|M.thermoresistible__bu VALGVLVLGERVGGLQRIAL-----------------------AVAAVAV
. . . .
MAB_3180|M.abscessus_ATCC_1997 NRLLATGGIDTVLVLLLVALLGLAAGGVYQQRFCADVDFRVNSTIQNLSG
MAV_0095|M.avium_104 QRLMSTHGVDPVIVLLFVALLGLSAGGVYQQRFCGGVDFRAMSALQNGVA
MSMEG_2542|M.smegmatis_MC2_155 SRLAAESGVDPVLLLLLAALLGLSAGGVYQQRFCSDVDFRAATAVQNAVA
MMAR_4903|M.marinum_M GGGGSQTDWLGVGLGLLAAVL-YAAGVLLQKVALRTVDALSATLWGTLVG
MUL_0503|M.ulcerans_Agy99 GGGGSQTDWLGVGLGLLAAVL-YAAGVLLQKVALRTVDALSATLWGTLVG
Mvan_5539|M.vanbaalenii_PYR-1 PELAGGSAWPVTEVLLVAVCYAIAP--LIAARYLAEVPAMPLTAACLGLA
Mflv_0187|M.gilvum_PYR-GCK EEVRDPARTIPRAIGLALVIALLTYAVVAVAVLAQLGSTGLATADAPLAD
TH_0580|M.thermoresistible__bu VVLTVEVGQPPYIALVLAASFGIYG----LVKKVVDADPRVGVAVETLLA
: .
MAB_3180|M.abscessus_ATCC_1997 LLPAGALALLTPSAVHDPWKGAAAVAAVVLLNA---TLCTS---------
MAV_0095|M.avium_104 LLPAAGLAMLTPFGVHDGRKALVAVAAMVLLNA---TLAVS---------
MSMEG_2542|M.smegmatis_MC2_155 LLPAAVFAALTPFAVHNAKHAVAAVAGVVLLNA---ALGVS---------
MMAR_4903|M.marinum_M FIVT--LPFLPAAARELSLASAADLWWLVFLGAGPSAIAFT---------
MUL_0503|M.ulcerans_Agy99 FIVT--LPFLPAAARELSLASAADLWWLVFLGAGPSAIAFT---------
Mvan_5539|M.vanbaalenii_PYR-1 ALIYVGPAAATWPAEIPSMRVLTAVALLAVVCT---SLAFI---------
Mflv_0187|M.gilvum_PYR-GCK AVRAAGLPALAPVVAVGAALAALGALLALLLGVSRTTLAMARDRHLPHPL
TH_0580|M.thermoresistible__bu LPLAGGFLIALHLLGEGSFGNHGAGHTVLLLAAG-PVTAIP---------
.: .
MAB_3180|M.abscessus_ATCC_1997 ---LYVRSVSTFGAAAVAMLFCVIPAVAGLLSWLLLGQ-------RVD-I
MAV_0095|M.avium_104 ---LYVRAINAHGAAAVAMLFAVIPAVAAVLSWIILGQ-------RPD-Q
MSMEG_2542|M.smegmatis_MC2_155 ---MYVRAISRHGAPAVTMLFCVIPAVAGVLSWLMLGE-------RPD-V
MMAR_4903|M.marinum_M ---TWAYALARTNAGVTAATTLMVPALVIALSGLLLAE-------VPNPL
MUL_0503|M.ulcerans_Agy99 ---TWAYALARTNAGVTAATTLMVPALVIALSGLLLAE-------VPNPL
Mvan_5539|M.vanbaalenii_PYR-1 ---LFFALIREVGAPRALVITYVNPAVALAAGVIVLNEP-LTARHLLG--
Mflv_0187|M.gilvum_PYR-GCK AHVHPGHGTPQRAEVAVGVVVAVVAAVVDVRSAIGFSSFGVLLYYAVANA
TH_0580|M.thermoresistible__bu -LLLFAAAAQRLPLVVLGLLFYLNPALQMAWGVLVNHES-----------
: .*: . : .
MAB_3180|M.abscessus_ATCC_1997 GMG------IGLVIGALACWLNARASRSTKLAVKPQSPS---IGAATAAP
MAV_0095|M.avium_104 GVA------AGLVLGGLACWVNTRAAS------RPRTQR---DGVPAAAP
MSMEG_2542|M.smegmatis_MC2_155 GIA------VGLVVGAAACWLNSSGSR------QQRQHD---PGRDG---
MMAR_4903|M.marinum_M GLAGGGACLIGVALARGLLRLPARSRRRTGTGDQLQGVESLPVGLSDYAA
MUL_0503|M.ulcerans_Agy99 GLAGGGACLIGVVLARGLLRLPARSRRRTGTGDQLQGVESLPVGLSDYAA
Mvan_5539|M.vanbaalenii_PYR-1 ---------LAMILAGSVLATRRPVEP------MQAAPR-----------
Mflv_0187|M.gilvum_PYR-GCK SAWTLTATPRARLVPAVGLIGCVVLAFTLPPVSVLAGAAVVLVGMLAYTT
TH_0580|M.thermoresistible__bu -------MPLGRWIGFALIWVALAVMAVDALRRGGAGTPNRPAAVPDRVV
. :
MAB_3180|M.abscessus_ATCC_1997 TREPAVAS--
MAV_0095|M.avium_104 AELQSVSR--
MSMEG_2542|M.smegmatis_MC2_155 ----------
MMAR_4903|M.marinum_M SSTAPSGRQL
MUL_0503|M.ulcerans_Agy99 SSTAPSGRQL
Mvan_5539|M.vanbaalenii_PYR-1 ----------
Mflv_0187|M.gilvum_PYR-GCK GGALRRGV--
TH_0580|M.thermoresistible__bu P---------