For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IADPAEYAGRLQRVHERMRTQSLSALVVCDPANIFYLTGYAAWSFYTPQCLVVPADGDPHLFLRAMDAAG AASLCNVAAERIHGYPEDLVHRPDTHPFRWIAAAVRSLIPAGEPVGVEGDANYFSPRAYFALAEGLPANR LVDAGELVNWVRLVKSPYEIGQLQIAGAIAEHAMRTACERIEPGRRQCDLVADILTAQTIGVDGHGGDFP AIVPMLPTGDAAGTPHCTWTDQPFLDGEATTIELAGVHGRYHAPLARTVMLGEPPQRLAETAKITAEAMQ ATLEAIRPGATGASVHRAFDDVIRPHGLRKDSRLGYSIGIGFPPDWGERTVSLRREDTAELAAGMAFHVI LGMWMDGWGYELSEPIVVTPTGFERLTNLPHELTIRR*
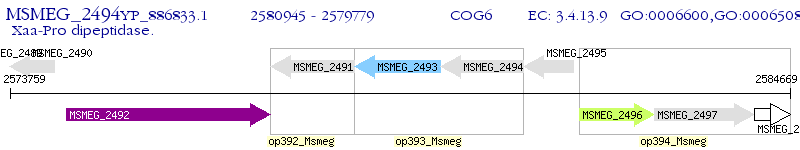
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2494 | - | - | 100% (388) | Xaa-Pro aminopeptidase |
| M. smegmatis MC2 155 | MSMEG_3881 | - | 2e-29 | 27.41% (394) | proline dipeptidase |
| M. smegmatis MC2 155 | MSMEG_3034 | - | 9e-17 | 26.29% (369) | metallopeptidase, M24 family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2116c | pepE | 1e-29 | 29.46% (387) | dipeptidase PepE |
| M. gilvum PYR-GCK | Mflv_3098 | - | 4e-30 | 27.66% (394) | peptidase M24 |
| M. tuberculosis H37Rv | Rv2089c | pepE | 1e-29 | 29.46% (387) | dipeptidase PepE |
| M. leprae Br4923 | MLBr_00521 | pepQ | 1e-13 | 27.27% (264) | putative cytoplasmic peptidase |
| M. abscessus ATCC 19977 | MAB_2192 | - | 3e-27 | 27.92% (394) | dipeptidase PepE |
| M. marinum M | MMAR_3075 | pepE | 1e-28 | 29.20% (387) | dipeptidase PepE |
| M. avium 104 | MAV_2414 | - | 8e-29 | 29.20% (387) | proline dipeptidase |
| M. thermoresistible (build 8) | TH_0446 | - | 2e-33 | 29.50% (383) | proline dipeptidase |
| M. ulcerans Agy99 | MUL_2321 | pepE | 3e-28 | 28.68% (387) | dipeptidase PepE |
| M. vanbaalenii PYR-1 | Mvan_3437 | - | 2e-29 | 28.68% (387) | peptidase M24 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3098|M.gilvum_PYR-GCK MTDARFGTDVYAHRLRAAAEAAATAGLSGLVITPGYDLRYLTGSRAQTFE
Mvan_3437|M.vanbaalenii_PYR-1 MSQDRFDTEVYARRLRAAAAAAADAGLAGLVITPGYDLRYLVGSRAQTFE
TH_0446|M.thermoresistible__bu MSANRFSTDVYATRLRAAAAATAEAGLSGLVITPGYDLQYLTGSRAQTFE
Mb2116c|M.bovis_AF2122/97 MGSRRFDAEVYARRLALAAAATADAGLAGLVITPGYDLCYLIGSRAETFE
Rv2089c|M.tuberculosis_H37Rv MGSRRFDAEVYARRLALAAAATADAGLAGLVITPGYDLCYLIGSRAETFE
MMAR_3075|M.marinum_M MDSGRFDTAVYARRLAAAAAATEQAGLAGLVITPGYDLRYLIGSRADTFE
MUL_2321|M.ulcerans_Agy99 MDSGRFDTAVYARRLAAAAAATEQAGLAGLVITPGYDLRYLIGSRADTFE
MAV_2414|M.avium_104 MESHRFDADVYARRLAAAASATAAAGLVGLVITPGYDLRYLVGSRAQTLE
MAB_2192|M.abscessus_ATCC_1997 MAPQRFESDVYADRLRRAAQEAEKVGVTGLVVTPGYDLRYLTGSRAQTFE
MSMEG_2494|M.smegmatis_MC2_155 ---MIADPAEYAGRLQRVHERMRTQSLSALVVCDPANIFYLTGYAAWSFY
MLBr_00521|M.leprae_Br4923 --------MTHSQRRDNLKSQIATTGLDAMLVTCLINVRYLSGFTGS---
:: * .: .::: :: ** * .
Mflv_3098|M.gilvum_PYR-GCK RLTALVLPAAGEPTIVVPRLELA---ALKESAIPELGVAVRDWVD--GEN
Mvan_3437|M.vanbaalenii_PYR-1 RLTALVLPATGDPTVVVPRLELA---SLKDSAVTELGVAVRDWVD--GDD
TH_0446|M.thermoresistible__bu RLMALVVPASGEPTLVLPRLELA---VLKDSAVPELGLTVRDWVD--GDD
Mb2116c|M.bovis_AF2122/97 RLTALVLPAAGAPAVVLPRLELA---ALKQSAAAELGLRVCDWVD--GDD
Rv2089c|M.tuberculosis_H37Rv RLTALVLPAAGAPAVVLPRLELA---ALKQSAAAELGLRVCDWVD--GDD
MMAR_3075|M.marinum_M RLTALVLPASGVPTIVLPRLELA---SLKESAASDLGVCVRDWVD--GDD
MUL_2321|M.ulcerans_Agy99 RLTALVLPASGVPTIVLPRLELA---SLKESAASDLGVCVRDWVD--GDD
MAV_2414|M.avium_104 RFTALVLPASGEPTVVVPRLEVA---SLKGSAIAELGLPVRDWVD--GEN
MAB_2192|M.abscessus_ATCC_1997 RLTALVVPATGQPTVVAPRMELA---ALRDSAIGDLNIPISDWVD--GEN
MSMEG_2494|M.smegmatis_MC2_155 TPQCLVVPADGDPHLFLRAMDAAGAASLCNVAAERIHGYPEDLVHRPDTH
MLBr_00521|M.leprae_Br4923 -NGALLVFADERGAVLATDGRYR---TQAAQQVPELEVAIEQSCG----R
.*:: * :. : :
Mflv_3098|M.gilvum_PYR-GCK PYQLVADALPGDGPQSVAVTDSMTALHLLP------LADVLGTVPVLATD
Mvan_3437|M.vanbaalenii_PYR-1 PYRLVVEALPGDGPLAVAVTDSMPALHLLP------LAEVLGAVPVLATD
TH_0446|M.thermoresistible__bu PYRLVAEALGAGPSGRPAVTDSMPALHLLP------LTDLLGSVPVLATS
Mb2116c|M.bovis_AF2122/97 PYGLVSAVLGG-APVATAVTDSMPALHMLP------LADALGVLPVLATD
Rv2089c|M.tuberculosis_H37Rv PYGLVSAVLGG-APVATAVTDSMPALHMLP------LADALGVLPVLATD
MMAR_3075|M.marinum_M PYQLVAEALGG-APAATAVTDSMPALHLLP------LADALGVLPVLATD
MUL_2321|M.ulcerans_Agy99 PYQLVAVALGG-APAATAVTDSMPALHLLP------LADALGVLPVLATD
MAV_2414|M.avium_104 PYEMVAAALGG-APAATAVTDSIPALHLLP------MAGVLGVLPVLATD
MAB_2192|M.abscessus_ATCC_1997 PYELVRAALGT--AGTVAVTESMPALHLLP------LAQLVGTVPVLATG
MSMEG_2494|M.smegmatis_MC2_155 PFRWIAAAVRSLIPAGEPVGVEGDANYFSPRAYFALAEGLPANRLVDAGE
MLBr_00521|M.leprae_Br4923 YLAGRAADDGVCILGFESHVVTVDGLDALTG-----ELEGRHTELVRAPG
. . . * *
Mflv_3098|M.gilvum_PYR-GCK VLRTLRMIKDPAEIDALRKAGAAIDRVHARVPEF--LVPGRTEADVAADI
Mvan_3437|M.vanbaalenii_PYR-1 VLRNLRMVKDPAEIDALRKAGAAIDRVHERVPQF--LRPGRTEADVAADI
TH_0446|M.thermoresistible__bu VLRRLRMIKDPAEIDALRKAGAAIDRVHARVPEF--LVPGRTEADVAADI
Mb2116c|M.bovis_AF2122/97 VLRRLRMVKEETEIDALRKAGAAIDRVHARVPEF--LVPGRTEADVAADI
Rv2089c|M.tuberculosis_H37Rv VLRRLRMVKEETEIDALRKAGAAIDRVHARVPEF--LVPGRTEADVAADI
MMAR_3075|M.marinum_M VLRQLRMVKEAAEVDALAKAGAAIDRVHARVPAF--LVPGRTEAQVAADI
MUL_2321|M.ulcerans_Agy99 VLRQLRMVKEAAEVDALAKAGAAIDRVHARVPAF--LVPGRTEAQVAADI
MAV_2414|M.avium_104 VLRGLRMVKEECEVDALRKAGAAIDRVHARVPSF--LVPGRTEADVAADI
MAB_2192|M.abscessus_ATCC_1997 VLRRLRMVKDASEIDALRKAGAAIDRVHALVPEL--LIPGRTEAQVAADI
MSMEG_2494|M.smegmatis_MC2_155 LVNWVRLVKSPYEIGQLQIAGAIAEHAMRTACER--IEPGRRQCDLVADI
MLBr_00521|M.leprae_Br4923 TVEVLREVKDAGELALLRLAAEAADAALTELVARGGVRPGLTEREVGREL
:. :* :*. *: * *. : . : ** : :: ::
Mflv_3098|M.gilvum_PYR-GCK A--QAIVAEGHS---EVAFIIVGSGPHGADPHHECSDRELQAGDIVVVDI
Mvan_3437|M.vanbaalenii_PYR-1 A--EAIVAEGHS---EVAFIIVGSGPHGADPHHECSDRELRAGDIVVVDI
TH_0446|M.thermoresistible__bu A--EAIVAEGHS---DVAFIIVGSGPHGADPHHECSDRTLQVGDIVVVDI
Mb2116c|M.bovis_AF2122/97 A--EAIVAEGHS---EVAFVIVGSGPHGADPHHGYSDRELREGDIVVVDI
Rv2089c|M.tuberculosis_H37Rv A--EAIVAEGHS---EVAFVIVGSGPHGADPHHGYSDRELREGDIVVVDI
MMAR_3075|M.marinum_M A--EAIVAEGHS---GVAFVIVGSGPHGADPHHGYSDRKLQVGDIVVVDI
MUL_2321|M.ulcerans_Agy99 A--EAIVAEGHS---AVAFVIVGSGPHGADPHHGYSDRKLQVGDIVVVDI
MAV_2414|M.avium_104 A--EAIVAEGHS---EVAFIIVGSGPHGADPHHGYSDRELQVGDIVVVDI
MAB_2192|M.abscessus_ATCC_1997 A--EAIVAEGHS---EVAFIIVGSGPHGADPHHECSDRELRTGDIVVVDI
MSMEG_2494|M.smegmatis_MC2_155 LTAQTIGVDGHGGDFPAIVPMLPTGDAAGTPHCTWTDQPFLDGEATTIEL
MLBr_00521|M.leprae_Br4923 EG----LMLDHGADALSFETIVAAGPNSAIPHHRPTDTVLAAGDFVKIDF
.*. :: :* .. ** :* : *: . :::
Mflv_3098|M.gilvum_PYR-GCK GGPYDPGYNSDSTRTYSIGEPDPEVARRYAVLQRAQRAAVDAVRPGVTAE
Mvan_3437|M.vanbaalenii_PYR-1 GGPYDPGYNSDSTRTYSIGEPDPEVARRYAVLQRAQRVAVDMVRPGVTAE
TH_0446|M.thermoresistible__bu GGPYAPGYHSDCTRTYSIGEPDPEIARRYAVLQRAQRAAVDAVRPGVTAE
Mb2116c|M.bovis_AF2122/97 GGTYGPGYHSDSTRTYSIGEPDSDVAQSYSMLQRAQRAAFEAIRPGVTAE
Rv2089c|M.tuberculosis_H37Rv GGTYGPGYHSDSTRTYSIGEPDSDVAQSYSMLQRAQRAAFEAIRPGVTAE
MMAR_3075|M.marinum_M GGTYEPGYYSDSTRTYSIGDPSPDVAQQYSALQRAQRAAVDAVRPGVTAA
MUL_2321|M.ulcerans_Agy99 GGTYEPGYYSDSTRTYSIGDPSPDVAQQYSALQRAQRAAVDAVRPGVTAA
MAV_2414|M.avium_104 GGSYEPGYHSDSTRTYSIGEPSHEVAQQYSILQRAQRAACDAVRPGVTAE
MAB_2192|M.abscessus_ATCC_1997 GGSYEPGYNSDSTRTYSIGEPDPDVAQRISVLERAQQAAVHAARPGVTAE
MSMEG_2494|M.smegmatis_MC2_155 AGVHGR-YHAPLARTVMLGEPPQRLAETAKITAEAMQATLEAIRPGATGA
MLBr_00521|M.leprae_Br4923 G-ALVAGYHSDMTRTFVLGPAADWQLEIYQLVADAQRAGRKALRPGANLR
. * : :** :* . . * :. . ***..
Mflv_3098|M.gilvum_PYR-GCK QVDAAARDVLADEGLADAFVHRTGHGIGLSVHEEPYIVAG---NALPLQE
Mvan_3437|M.vanbaalenii_PYR-1 QVDAAARDVLAAEGLAEAFVHRTGHGIGLSVHEEPYIVAG---NSLPLQE
TH_0446|M.thermoresistible__bu QIDAAARDVLAAEGLAEAFVHRTGHGIGLSVHEEPYIVAG---NNVPLEA
Mb2116c|M.bovis_AF2122/97 QVDAAARDVLAEAGLAEYFVHRTGHGIGLCVHEEPYIVAG---NDLVLVP
Rv2089c|M.tuberculosis_H37Rv QVDAAARDVLAEAGLAEYFVHRTGHGIGLCVHEEPYIVAG---NDLVLVP
MMAR_3075|M.marinum_M QVDAAARDVLADAGLAEYFVHRTGHGIGLCVHEEPYIVAG---NELPLVA
MUL_2321|M.ulcerans_Agy99 QVDAAARDVLADAGLAEYFVHRTGHGIGLCVHEEPYIVAG---NELPLVA
MAV_2414|M.avium_104 QVDAAARDVLAAAGLAEYFVHRTGHGIGLSVHEEPYIVAG---NDLPLAA
MAB_2192|M.abscessus_ATCC_1997 SVDAAARRVLTEAGMGEAFVHRTGHGIGLSVHEEPYIVEG---NELVLEP
MSMEG_2494|M.smegmatis_MC2_155 SVHRAFDDVIRPHGLRKDSRLGYSIGIGFPPDWGERTVSLRREDTAELAA
MLBr_00521|M.leprae_Br4923 DVDAAARQLIVDAGYGQQFSHGLGHGVGLEIHEAPGIGTT---SAGTLLA
.:. * :: * . . *:*: . . *
Mflv_3098|M.gilvum_PYR-GCK GMAFSVEPGIYFPGQWGARIEDIVVVTADG--ALAVNNRPHDLVVVPVP-
Mvan_3437|M.vanbaalenii_PYR-1 GMAFSVEPGIYFPGQWGARIEDIVIVTGDG--AEPVNHRPHELVVVPVP-
TH_0446|M.thermoresistible__bu GMAFSVEPGIYFPGQWGARIEDIVIVTADG--AEPVNHRPHELVVVVPV-
Mb2116c|M.bovis_AF2122/97 GMAFSIEPGIYFPGRWGARIEDIVIVTEDG--AVSVNNCPHELIVVPVS-
Rv2089c|M.tuberculosis_H37Rv GMAFSIEPGIYFPGRWGARIEDIVIVTEDG--AVSVNNCPHELIVVPVS-
MMAR_3075|M.marinum_M GMAFSVEPGIYFPGRWGARIEDIVVVTEDG--ALSVNNRPHELMVVPVS-
MUL_2321|M.ulcerans_Agy99 GMAFSIEPGIYFPGRWGARIEDIVVVTENG--ALSVNNRPHELMVVPVS-
MAV_2414|M.avium_104 GMAFSIEPGIYFPGRWGARIEDIVVVTDDG--ALAVNNRPHELIVVPVP-
MAB_2192|M.abscessus_ATCC_1997 GMAFSIEPGVYFPGQWGARIEDIVVITENG--CESVNSRPHGLTVVPTR-
MSMEG_2494|M.smegmatis_MC2_155 GMAFHVILGMWMDG-WGYELSEPIVVTPTG--FERLTNLPHELTIRR---
MLBr_00521|M.leprae_Br4923 GSVVTVEPGVYLSGRGGVRIEDTLVVAAKGTSETPITSRPTGFIPELLTR
* .. : *::: * * .:.: :::: * :. * :
Mflv_3098|M.gilvum_PYR-GCK --------
Mvan_3437|M.vanbaalenii_PYR-1 --------
TH_0446|M.thermoresistible__bu --------
Mb2116c|M.bovis_AF2122/97 --------
Rv2089c|M.tuberculosis_H37Rv --------
MMAR_3075|M.marinum_M --------
MUL_2321|M.ulcerans_Agy99 --------
MAV_2414|M.avium_104 --------
MAB_2192|M.abscessus_ATCC_1997 --------
MSMEG_2494|M.smegmatis_MC2_155 --------
MLBr_00521|M.leprae_Br4923 FPKELAIL