For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAPQRFESDVYADRLRRAAQEAEKVGVTGLVVTPGYDLRYLTGSRAQTFERLTALVVPATGQPTVVAPRM ELAALRDSAIGDLNIPISDWVDGENPYELVRAALGTAGTVAVTESMPALHLLPLAQLVGTVPVLATGVLR RLRMVKDASEIDALRKAGAAIDRVHALVPELLIPGRTEAQVAADIAEAIVAEGHSEVAFIIVGSGPHGAD PHHECSDRELRTGDIVVVDIGGSYEPGYNSDSTRTYSIGEPDPDVAQRISVLERAQQAAVHAARPGVTAE SVDAAARRVLTEAGMGEAFVHRTGHGIGLSVHEEPYIVEGNELVLEPGMAFSIEPGVYFPGQWGARIEDI VVITENGCESVNSRPHGLTVVPTR
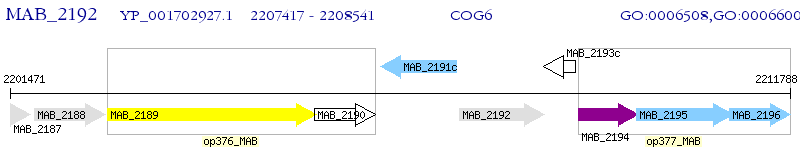
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2192 | - | - | 100% (374) | dipeptidase PepE |
| M. abscessus ATCC 19977 | MAB_2838c | - | 5e-39 | 40.50% (242) | putative cytoplasmic peptidase PepQ |
| M. abscessus ATCC 19977 | MAB_0094c | - | 6e-12 | 26.02% (246) | methionine aminopeptidase, type I (MAP) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2116c | pepE | 1e-155 | 73.46% (373) | dipeptidase PepE |
| M. gilvum PYR-GCK | Mflv_3098 | - | 1e-164 | 75.94% (374) | peptidase M24 |
| M. tuberculosis H37Rv | Rv2089c | pepE | 1e-155 | 73.46% (373) | dipeptidase PepE |
| M. leprae Br4923 | MLBr_00521 | pepQ | 2e-39 | 32.71% (373) | putative cytoplasmic peptidase |
| M. marinum M | MMAR_3075 | pepE | 1e-154 | 73.17% (369) | dipeptidase PepE |
| M. avium 104 | MAV_2414 | - | 1e-156 | 73.19% (373) | proline dipeptidase |
| M. smegmatis MC2 155 | MSMEG_3881 | - | 1e-164 | 76.00% (375) | proline dipeptidase |
| M. thermoresistible (build 8) | TH_0446 | - | 1e-160 | 73.73% (373) | proline dipeptidase |
| M. ulcerans Agy99 | MUL_2321 | pepE | 1e-155 | 73.71% (369) | dipeptidase PepE |
| M. vanbaalenii PYR-1 | Mvan_3437 | - | 1e-161 | 75.13% (374) | peptidase M24 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3098|M.gilvum_PYR-GCK MTDARFGTDVYAHRLRAAAEAAATAGLSGLVITPGYDLRYLTGSRAQTFE
Mvan_3437|M.vanbaalenii_PYR-1 MSQDRFDTEVYARRLRAAAAAAADAGLAGLVITPGYDLRYLVGSRAQTFE
MSMEG_3881|M.smegmatis_MC2_155 MTISRFSTDVYAQRLQTAAQAAGDAGLAGLVITPGYDLRYLVGSRAQTFE
TH_0446|M.thermoresistible__bu MSANRFSTDVYATRLRAAAAATAEAGLSGLVITPGYDLQYLTGSRAQTFE
Mb2116c|M.bovis_AF2122/97 MGSRRFDAEVYARRLALAAAATADAGLAGLVITPGYDLCYLIGSRAETFE
Rv2089c|M.tuberculosis_H37Rv MGSRRFDAEVYARRLALAAAATADAGLAGLVITPGYDLCYLIGSRAETFE
MMAR_3075|M.marinum_M MDSGRFDTAVYARRLAAAAAATEQAGLAGLVITPGYDLRYLIGSRADTFE
MUL_2321|M.ulcerans_Agy99 MDSGRFDTAVYARRLAAAAAATEQAGLAGLVITPGYDLRYLIGSRADTFE
MAV_2414|M.avium_104 MESHRFDADVYARRLAAAASATAAAGLVGLVITPGYDLRYLVGSRAQTLE
MAB_2192|M.abscessus_ATCC_1997 MAPQRFESDVYADRLRRAAQEAEKVGVTGLVVTPGYDLRYLTGSRAQTFE
MLBr_00521|M.leprae_Br4923 --------MTHSQRRDNLKSQIATTGLDAMLVTCLINVRYLSGFTGS---
.:: * .*: .:::* :: ** * ..
Mflv_3098|M.gilvum_PYR-GCK RLTALVLPAAGEPTIVVPRLELAALKESAIPELGVAVRDWVDGENPYQLV
Mvan_3437|M.vanbaalenii_PYR-1 RLTALVLPATGDPTVVVPRLELASLKDSAVTELGVAVRDWVDGDDPYRLV
MSMEG_3881|M.smegmatis_MC2_155 RLTALVLPADGDPTIVVPRLELASLKESAVTDLGVAVRDWVDGENPYAMV
TH_0446|M.thermoresistible__bu RLMALVVPASGEPTLVLPRLELAVLKDSAVPELGLTVRDWVDGDDPYRLV
Mb2116c|M.bovis_AF2122/97 RLTALVLPAAGAPAVVLPRLELAALKQSAAAELGLRVCDWVDGDDPYGLV
Rv2089c|M.tuberculosis_H37Rv RLTALVLPAAGAPAVVLPRLELAALKQSAAAELGLRVCDWVDGDDPYGLV
MMAR_3075|M.marinum_M RLTALVLPASGVPTIVLPRLELASLKESAASDLGVCVRDWVDGDDPYQLV
MUL_2321|M.ulcerans_Agy99 RLTALVLPASGVPTIVLPRLELASLKESAASDLGVCVRDWVDGDDPYQLV
MAV_2414|M.avium_104 RFTALVLPASGEPTVVVPRLEVASLKGSAIAELGLPVRDWVDGENPYEMV
MAB_2192|M.abscessus_ATCC_1997 RLTALVVPATGQPTVVAPRMELAALRDSAIGDLNIPISDWVDGENPYELV
MLBr_00521|M.leprae_Br4923 -NGALLVFADERGAVLATDGRYRTQAAQQVPELEVAIEQSCGRYLAGRAA
**:: * ::: . . . :* : : : . . .
Mflv_3098|M.gilvum_PYR-GCK ADALPGDGPQSVAVTDSMTALHLLPLADVLGTVPVLATDVLRTLRMIKDP
Mvan_3437|M.vanbaalenii_PYR-1 VEALPGDGPLAVAVTDSMPALHLLPLAEVLGAVPVLATDVLRNLRMVKDP
MSMEG_3881|M.smegmatis_MC2_155 VEALGGPG-RNAAVTDAMPALHLLPLADLLGVVPVLATDVLRRLRMIKDD
TH_0446|M.thermoresistible__bu AEALGAGPSGRPAVTDSMPALHLLPLTDLLGSVPVLATSVLRRLRMIKDP
Mb2116c|M.bovis_AF2122/97 SAVLGGAP-VATAVTDSMPALHMLPLADALGVLPVLATDVLRRLRMVKEE
Rv2089c|M.tuberculosis_H37Rv SAVLGGAP-VATAVTDSMPALHMLPLADALGVLPVLATDVLRRLRMVKEE
MMAR_3075|M.marinum_M AEALGGAP-AATAVTDSMPALHLLPLADALGVLPVLATDVLRQLRMVKEA
MUL_2321|M.ulcerans_Agy99 AVALGGAP-AATAVTDSMPALHLLPLADALGVLPVLATDVLRQLRMVKEA
MAV_2414|M.avium_104 AAALGGAP-AATAVTDSIPALHLLPMAGVLGVLPVLATDVLRGLRMVKEE
MAB_2192|M.abscessus_ATCC_1997 RAALGTAG--TVAVTESMPALHLLPLAQLVGTVPVLATGVLRRLRMVKDA
MLBr_00521|M.leprae_Br4923 DDGVCILG-FESHVVTVDGLDALTGELEGRHTELVRAPGTVEVLREVKDA
: *. : * *...:. ** :*:
Mflv_3098|M.gilvum_PYR-GCK AEIDALRKAGAAIDRVHARVPEF--LVPGRTEADVAADIAQAIVAEGHSE
Mvan_3437|M.vanbaalenii_PYR-1 AEIDALRKAGAAIDRVHERVPQF--LRPGRTEADVAADIAEAIVAEGHSE
MSMEG_3881|M.smegmatis_MC2_155 AEIDALRKAGAAIDRVHARVPEF--LVPGRTEADVAADIAEAIVAEGHSE
TH_0446|M.thermoresistible__bu AEIDALRKAGAAIDRVHARVPEF--LVPGRTEADVAADIAEAIVAEGHSD
Mb2116c|M.bovis_AF2122/97 TEIDALRKAGAAIDRVHARVPEF--LVPGRTEADVAADIAEAIVAEGHSE
Rv2089c|M.tuberculosis_H37Rv TEIDALRKAGAAIDRVHARVPEF--LVPGRTEADVAADIAEAIVAEGHSE
MMAR_3075|M.marinum_M AEVDALAKAGAAIDRVHARVPAF--LVPGRTEAQVAADIAEAIVAEGHSG
MUL_2321|M.ulcerans_Agy99 AEVDALAKAGAAIDRVHARVPAF--LVPGRTEAQVAADIAEAIVAEGHSA
MAV_2414|M.avium_104 CEVDALRKAGAAIDRVHARVPSF--LVPGRTEADVAADIAEAIVAEGHSE
MAB_2192|M.abscessus_ATCC_1997 SEIDALRKAGAAIDRVHALVPEL--LIPGRTEAQVAADIAEAIVAEGHSE
MLBr_00521|M.leprae_Br4923 GELALLRLAAEAADAALTELVARGGVRPGLTEREVGRELEGLMLDHGADA
*: * *. * * . : : ** ** :*. :: :: .* .
Mflv_3098|M.gilvum_PYR-GCK VAFI-IVGSGPHGADPHHECSDRELQAGDIVVVDIGGPYDPGYNSDSTRT
Mvan_3437|M.vanbaalenii_PYR-1 VAFI-IVGSGPHGADPHHECSDRELRAGDIVVVDIGGPYDPGYNSDSTRT
MSMEG_3881|M.smegmatis_MC2_155 AAFI-IVGSGPNGADPHHECSDRELRVGDIVVVDIGGPYEPGYNSDCTRT
TH_0446|M.thermoresistible__bu VAFI-IVGSGPHGADPHHECSDRTLQVGDIVVVDIGGPYAPGYHSDCTRT
Mb2116c|M.bovis_AF2122/97 VAFV-IVGSGPHGADPHHGYSDRELREGDIVVVDIGGTYGPGYHSDSTRT
Rv2089c|M.tuberculosis_H37Rv VAFV-IVGSGPHGADPHHGYSDRELREGDIVVVDIGGTYGPGYHSDSTRT
MMAR_3075|M.marinum_M VAFV-IVGSGPHGADPHHGYSDRKLQVGDIVVVDIGGTYEPGYYSDSTRT
MUL_2321|M.ulcerans_Agy99 VAFV-IVGSGPHGADPHHGYSDRKLQVGDIVVVDIGGTYEPGYYSDSTRT
MAV_2414|M.avium_104 VAFI-IVGSGPHGADPHHGYSDRELQVGDIVVVDIGGSYEPGYHSDSTRT
MAB_2192|M.abscessus_ATCC_1997 VAFI-IVGSGPHGADPHHECSDRELRTGDIVVVDIGGSYEPGYNSDSTRT
MLBr_00521|M.leprae_Br4923 LSFETIVAAGPNSAIPHHRPTDTVLAAGDFVKIDFG-ALVAGYHSDMTRT
:* **.:**:.* *** :* * **:* :*:* . .** ** ***
Mflv_3098|M.gilvum_PYR-GCK YSIGEPDPEVARRYAVLQRAQRAAVDAVRPGVTAEQVDAAARDVLADEGL
Mvan_3437|M.vanbaalenii_PYR-1 YSIGEPDPEVARRYAVLQRAQRVAVDMVRPGVTAEQVDAAARDVLAAEGL
MSMEG_3881|M.smegmatis_MC2_155 YSIGEPDPEVARRYALLQQAQQAAVAAVRPGVTAEQVDAAARDVLAAEGL
TH_0446|M.thermoresistible__bu YSIGEPDPEIARRYAVLQRAQRAAVDAVRPGVTAEQIDAAARDVLAAEGL
Mb2116c|M.bovis_AF2122/97 YSIGEPDSDVAQSYSMLQRAQRAAFEAIRPGVTAEQVDAAARDVLAEAGL
Rv2089c|M.tuberculosis_H37Rv YSIGEPDSDVAQSYSMLQRAQRAAFEAIRPGVTAEQVDAAARDVLAEAGL
MMAR_3075|M.marinum_M YSIGDPSPDVAQQYSALQRAQRAAVDAVRPGVTAAQVDAAARDVLADAGL
MUL_2321|M.ulcerans_Agy99 YSIGDPSPDVAQQYSALQRAQRAAVDAVRPGVTAAQVDAAARDVLADAGL
MAV_2414|M.avium_104 YSIGEPSHEVAQQYSILQRAQRAACDAVRPGVTAEQVDAAARDVLAAAGL
MAB_2192|M.abscessus_ATCC_1997 YSIGEPDPDVAQRISVLERAQQAAVHAARPGVTAESVDAAARRVLTEAGM
MLBr_00521|M.leprae_Br4923 FVLGPAADWQLEIYQLVADAQRAGRKALRPGANLRDVDAAARQLIVDAGY
: :* . . : **:.. ***.. .:***** ::. *
Mflv_3098|M.gilvum_PYR-GCK ADAFVHRTGHGIGLSVHEEPYIVAGNALPLQEGMAFSVEPGIYFPGQWGA
Mvan_3437|M.vanbaalenii_PYR-1 AEAFVHRTGHGIGLSVHEEPYIVAGNSLPLQEGMAFSVEPGIYFPGQWGA
MSMEG_3881|M.smegmatis_MC2_155 AEAFVHRTGHGIGLSVHEEPYIVAGNNLPLERGMAFSVEPGVYFPGQWGA
TH_0446|M.thermoresistible__bu AEAFVHRTGHGIGLSVHEEPYIVAGNNVPLEAGMAFSVEPGIYFPGQWGA
Mb2116c|M.bovis_AF2122/97 AEYFVHRTGHGIGLCVHEEPYIVAGNDLVLVPGMAFSIEPGIYFPGRWGA
Rv2089c|M.tuberculosis_H37Rv AEYFVHRTGHGIGLCVHEEPYIVAGNDLVLVPGMAFSIEPGIYFPGRWGA
MMAR_3075|M.marinum_M AEYFVHRTGHGIGLCVHEEPYIVAGNELPLVAGMAFSVEPGIYFPGRWGA
MUL_2321|M.ulcerans_Agy99 AEYFVHRTGHGIGLCVHEEPYIVAGNELPLVAGMAFSIEPGIYFPGRWGA
MAV_2414|M.avium_104 AEYFVHRTGHGIGLSVHEEPYIVAGNDLPLAAGMAFSIEPGIYFPGRWGA
MAB_2192|M.abscessus_ATCC_1997 GEAFVHRTGHGIGLSVHEEPYIVEGNELVLEPGMAFSIEPGVYFPGQWGA
MLBr_00521|M.leprae_Br4923 GQQFSHGLGHGVGLEIHEAPGIGTTSAGTLLAGSVVTVEPGVYLSGRGGV
.: * * ***:** :** * * . * * ..::***:*:.*: *.
Mflv_3098|M.gilvum_PYR-GCK RIEDIVVVTADG--ALAVNNRPHDLVVVPVP---------
Mvan_3437|M.vanbaalenii_PYR-1 RIEDIVIVTGDG--AEPVNHRPHELVVVPVP---------
MSMEG_3881|M.smegmatis_MC2_155 RIEDIVIVTEDG--AQSVNNQPHDLVVVPVR---------
TH_0446|M.thermoresistible__bu RIEDIVIVTADG--AEPVNHRPHELVVVVPV---------
Mb2116c|M.bovis_AF2122/97 RIEDIVIVTEDG--AVSVNNCPHELIVVPVS---------
Rv2089c|M.tuberculosis_H37Rv RIEDIVIVTEDG--AVSVNNCPHELIVVPVS---------
MMAR_3075|M.marinum_M RIEDIVVVTEDG--ALSVNNRPHELMVVPVS---------
MUL_2321|M.ulcerans_Agy99 RIEDIVVVTENG--ALSVNNRPHELMVVPVS---------
MAV_2414|M.avium_104 RIEDIVVVTDDG--ALAVNNRPHELIVVPVP---------
MAB_2192|M.abscessus_ATCC_1997 RIEDIVVITENG--CESVNSRPHGLTVVPTR---------
MLBr_00521|M.leprae_Br4923 RIEDTLVVAAKGTSETPITSRPTGFIPELLTRFPKELAIL
**** :::: .* .:. * :