For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTISQRRERLRQRLAAADLDAMLVTDLVNVRYLSGFTGSNAALLVRVDDATPVLATDGRYRTQAAQQAPD AEVVIERACGPHLAARAAADGVRRLGFESHVVTVEAHSALSEAAGDKVELVRAAGTVEALREVKDAGEIA MLRLACEAADAALTDLVERGGLRPGRTEKEVRRDLESLMLDHGADGPSFETIVATGANSAIPHHRPTDAV LATGDFVKIDFGALVSGYHSDMTRTFILGRAEQWQLDLYELVAASQAAGREALAAGVELRAVDAASRQVI IDAGYGDHFNHGLGHGVGLQIHEAPGLNSAAAGTLLAGSAVTVEPGVYLPGRGGVRIEDTLVVPGGESAE GVVDRFAHNISDAAPELLTRFPKELAIV
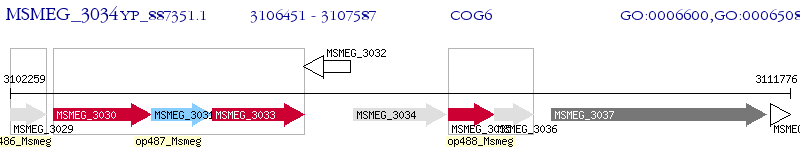
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3034 | - | - | 100% (378) | metallopeptidase, M24 family protein |
| M. smegmatis MC2 155 | MSMEG_3881 | - | 1e-34 | 32.47% (348) | proline dipeptidase |
| M. smegmatis MC2 155 | MSMEG_5683 | map | 5e-20 | 30.65% (261) | methionine aminopeptidase, type I |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2564c | pepQ | 1e-152 | 72.30% (379) | putative cytoplasmic peptidase PEPQ |
| M. gilvum PYR-GCK | Mflv_3753 | - | 1e-163 | 78.84% (378) | peptidase M24 |
| M. tuberculosis H37Rv | Rv2535c | pepQ | 1e-152 | 72.30% (379) | cytoplasmic peptidase PepQ |
| M. leprae Br4923 | MLBr_00521 | pepQ | 1e-145 | 69.92% (379) | putative cytoplasmic peptidase |
| M. abscessus ATCC 19977 | MAB_2838c | - | 1e-126 | 65.14% (370) | putative cytoplasmic peptidase PepQ |
| M. marinum M | MMAR_2180 | pepQ | 1e-153 | 73.09% (379) | cytoplasmic peptidase PepQ |
| M. avium 104 | MAV_3412 | - | 1e-149 | 72.44% (381) | peptidase, M24 family protein |
| M. thermoresistible (build 8) | TH_0106 | pepQ | 1e-153 | 77.87% (357) | PROBABLE CYTOPLASMIC PEPTIDASE PEPQ |
| M. ulcerans Agy99 | MUL_1764 | pepQ | 1e-154 | 73.09% (379) | cytoplasmic peptidase PepQ |
| M. vanbaalenii PYR-1 | Mvan_2650 | - | 1e-161 | 76.98% (378) | peptidase M24 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3753|M.gilvum_PYR-GCK MTISARRDRLRRLLADAGLDAMLVSDLVNVRYLSGFTGSNAALLVLAQD-
Mvan_2650|M.vanbaalenii_PYR-1 MTISQRRDRLRQRLADAELDAMLVSDLVNVRYLSGFTGSNAALLILAED-
MSMEG_3034|M.smegmatis_MC2_155 MTISQRRERLRQRLAAADLDAMLVTDLVNVRYLSGFTGSNAALLVRVDD-
TH_0106|M.thermoresistible__bu ---------------------MLVTDLTNVRYLSGFTGSNAALLVKADG-
MMAR_2180|M.marinum_M MTHSQRRHNLKSKISAAGLDAMLVTDLINVRYLSGFSGSNGALLVFADE-
MUL_1764|M.ulcerans_Agy99 MTHSQRRHNLKSKISAAGLDAMLVTDLINVRYLSGFSGSNGALLVFADE-
Mb2564c|M.bovis_AF2122/97 MTHSQRRDKLKAQIAASGLDAMLISDLINVRYLSGFSGSNGALLVFADE-
Rv2535c|M.tuberculosis_H37Rv MTHSQRRDKLKAQIAASGLDAMLISDLINVRYLSGFSGSNGALLVFADE-
MLBr_00521|M.leprae_Br4923 MTHSQRRDNLKSQIATTGLDAMLVTCLINVRYLSGFTGSNGALLVFADE-
MAV_3412|M.avium_104 MTHSQRRDRLRARLEADGLDALLVTDLINVRYLSGFTGSNGALLVFADD-
MAB_2838c|M.abscessus_ATCC_199 ------------MLRAEDLDALLVTDLINIRYLTGFTGSAAALWVAAGDG
:*:: * *:***:**:** .** : .
Mflv_3753|M.gilvum_PYR-GCK ---ETPVLATDGRYRTQAAHQAPDAEVVIERACGPHLAARAAADGVRRLG
Mvan_2650|M.vanbaalenii_PYR-1 ---ETPVLATDGRYRTQAAHQAPDAEVVIERACGPRLAARAAAAGIRRLG
MSMEG_3034|M.smegmatis_MC2_155 ---ATPVLATDGRYRTQAAQQAPDAEVVIERACGPHLAARAAADGVRRLG
TH_0106|M.thermoresistible__bu ---ATAVLATDGRYRTQAARQAPDVELVIERACGPHLAGRAAADGIRRLG
MMAR_2180|M.marinum_M ---REAVLATDGRYRTQAAHQAPDLEVAIERAIGRHLSCRAADDGVGRLG
MUL_1764|M.ulcerans_Agy99 ---REAVLATDGRYRTQAAHQAPDLEVAIERAIGRHLSCRAADDGVGRLG
Mb2564c|M.bovis_AF2122/97 ---RDAVLATDGRYRTQAASQAPDLEVAIERAVGRYLAGRAGEAGVGKLG
Rv2535c|M.tuberculosis_H37Rv ---RDAVLATDGRYRTQAASQAPDLEVAIERAVGRYLAGRAGEAGVGKLG
MLBr_00521|M.leprae_Br4923 ---RGAVLATDGRYRTQAAQQVPELEVAIEQSCGRYLAGRAADDGVCILG
MAV_3412|M.avium_104 ---RPPVLATDGRYRTQAAEQAPDLDVAIERALGRYLAGRAAEAGARTLG
MAB_2838c|M.abscessus_ATCC_199 PEEKATVLCTDGRYRTQAAAQVPDLRALIDRRYGAALVAHAPAG---RAG
.**.********** *.*: *:: * * :* *
Mflv_3753|M.gilvum_PYR-GCK FESHVVTVDAFTALS---EAAGNGCEFVRAAGTVEGLREVKDAGEVALLR
Mvan_2650|M.vanbaalenii_PYR-1 FESHVVTVDAFTALT---KAVGEACEFVRAAGTVEGLREVKDAGEIALLR
MSMEG_3034|M.smegmatis_MC2_155 FESHVVTVEAHSALS---EAAGDKVELVRAAGTVEALREVKDAGEIAMLR
TH_0106|M.thermoresistible__bu FESHVVTVDAHTALQ---QAA-EGVELVRAPRIVERLREVKDAGEIAILR
MMAR_2180|M.marinum_M FESHVVTVDGFDALTGALADT--NTELVRASGTVEALREIKDAGELALLR
MUL_1764|M.ulcerans_Agy99 FESHVVTVDGFDALTGALADT--NTELVRASRTVEALREIKDAGELALLR
Mb2564c|M.bovis_AF2122/97 FESHVVTVDGLDALAGALEGK--NTELVRASGTVESLREVKDAGELALLR
Rv2535c|M.tuberculosis_H37Rv FESHVVTVDGLDALAGALEGK--NTELVRASGTVESLREVKDAGELALLR
MLBr_00521|M.leprae_Br4923 FESHVVTVDGLDALTGELEGR--HTELVRAPGTVEVLREVKDAGELALLR
MAV_3412|M.avium_104 FESNVVTVDGFDALTGELDGRKAATELVKAAGTVEALREVKDAGEVALLR
MAB_2838c|M.abscessus_ATCC_199 FESHVVTVDGFDKLA----EAATDVELVRAPNRIEKLREVKDDSEVLLLE
***:****:. * *:*:*. :* ***:** .*: :*.
Mflv_3753|M.gilvum_PYR-GCK LACEAADAALKDLLERGGLRAGRTERDVRNELESLMLAHGADGASFETIV
Mvan_2650|M.vanbaalenii_PYR-1 LACEAADAALRDLVDQGGLRAGRTEKEVCNELESLMLAHGADGASFETIV
MSMEG_3034|M.smegmatis_MC2_155 LACEAADAALTDLVERGGLRPGRTEKEVRRDLESLMLDHGADGPSFETIV
TH_0106|M.thermoresistible__bu LACEAADAALTDLVAGGGLRPGRTEKQVRRDLESLMLDHGADGPSFETIV
MMAR_2180|M.marinum_M LACEAADAALSDLIKRGGLRPGRTEREVARDLEGLMLDHGADAVSFETIV
MUL_1764|M.ulcerans_Agy99 LACEAADAALSDLIKRGGLRPGRTEREVARDLEGLMLDHGADAVSFETIV
Mb2564c|M.bovis_AF2122/97 LACEAADAALTDLVARGGLRPGRTERQVSRELEALMLDHGADAVSFETIV
Rv2535c|M.tuberculosis_H37Rv LACEAADAALTDLVARGGLRPGRTERQVSRELEALMLDHGADAVSFETIV
MLBr_00521|M.leprae_Br4923 LAAEAADAALTELVARGGVRPGLTEREVGRELEGLMLDHGADALSFETIV
MAV_3412|M.avium_104 LACEAADAALTALVDGGGLRPGRTEREVSRELEALMLDHGADGISFETIV
MAB_2838c|M.abscessus_ATCC_199 RACAAADTALAALIETGGIRPGRTEREVARDLENLMAEHGADGPSFETIV
*. ***:** *: **:*.* **::* .:** ** ****. ******
Mflv_3753|M.gilvum_PYR-GCK AAGANSAIPHHRPTDAVLAAGDFVKIDFGALVAGYHSDMTRTFVLAPVAD
Mvan_2650|M.vanbaalenii_PYR-1 ATGANSAIPHHRPTDAVLAAGDFVKIDFGALVAGYHSDMTRTFVLAPIAD
MSMEG_3034|M.smegmatis_MC2_155 ATGANSAIPHHRPTDAVLATGDFVKIDFGALVSGYHSDMTRTFILGRAEQ
TH_0106|M.thermoresistible__bu AAGANSAIPHHRPTDAELKAGDFVKIDFGALVAGYHSDMTRTFVLGPAAQ
MMAR_2180|M.marinum_M AAGPNSAIPHHRPTDAVLAKGDFVKIDFGALVAGYHSDMTRTFVLDKAAD
MUL_1764|M.ulcerans_Agy99 AAGPNSAIPHHRPTDAVLAKGDFVKIDFGALVAGYHSDMTRTFVLDKAAD
Mb2564c|M.bovis_AF2122/97 AAGANSAIPHHRPTDAVLQVGDFVKIDFGALVAGYHSDMTRTFVLGKAAD
Rv2535c|M.tuberculosis_H37Rv AAGANSAIPHHRPTDAVLQVGDFVKIDFGALVAGYHSDMTRTFVLGKAAD
MLBr_00521|M.leprae_Br4923 AAGPNSAIPHHRPTDTVLAAGDFVKIDFGALVAGYHSDMTRTFVLGPAAD
MAV_3412|M.avium_104 ATGANSAIPHHRPTDAVLADGDFVKIDFGALVGGYHSDMTRTFVLTKAAD
MAB_2838c|M.abscessus_ATCC_199 AAGPNSAIPHHRPTGAMLAAGDFVKIDFGALVAGYHSDMTRTLVLGSAAP
*:*.**********.: * ************.*********::*
Mflv_3753|M.gilvum_PYR-GCK WQREIYSLVAASQQAGRDALAPGVALSTVDAAARQVIADAGYAENFGHGL
Mvan_2650|M.vanbaalenii_PYR-1 WQRDIYDLVATAQRAGTDALEPGVALKDVDAASRQVIVEAGYGENFGHGL
MSMEG_3034|M.smegmatis_MC2_155 WQLDLYELVAASQAAGREALAAGVELRAVDAASRQVIIDAGYGDHFNHGL
TH_0106|M.thermoresistible__bu WQHELYELVATAQRAGTEALRAGAPLREVDAAARQVIADAGYADNFGHGL
MMAR_2180|M.marinum_M WQLEIYELVAAAQKAGREALSAGAELRDVDGAARQMIVAAGHGDNFGHGL
MUL_1764|M.ulcerans_Agy99 WQLEIYELVAAAQKAGREALSAGAELRDVDGAARQMIVDAGHGDNFGHGL
Mb2564c|M.bovis_AF2122/97 WQLEIYQLVAEAQQAGRQALLPGAELRGVDAAARQLIADAGYGEHFGHGL
Rv2535c|M.tuberculosis_H37Rv WQLEIYQLVAEAQQAGRQALLPGAELRGVDAAARQLIADAGYGEHFGHGL
MLBr_00521|M.leprae_Br4923 WQLEIYQLVADAQRAGRKALRPGANLRDVDAAARQLIVDAGYGQQFSHGL
MAV_3412|M.avium_104 WQLEIYRLVADAQRAGREALRAGADLREVDAAARRVIADAGYGEQFSHSL
MAB_2838c|M.abscessus_ATCC_199 WQQEIHELVLTAQAAGRAALAPGASLKNVDAAARDVITAAGYGENFGHGL
** ::: ** :* ** ** .*. * **.*:* :* **:.::*.*.*
Mflv_3753|M.gilvum_PYR-GCK GHGVGLQIHEAPGINSAAAGTLLAGSAVTVEPGVYLPDRGGVRIEDTLVV
Mvan_2650|M.vanbaalenii_PYR-1 GHGVGLQIHEAPGINAAAAGTLLAGSVVTVEPGVYLPDRGGVRIEDTLVV
MSMEG_3034|M.smegmatis_MC2_155 GHGVGLQIHEAPGLNSAAAGTLLAGSAVTVEPGVYLPGRGGVRIEDTLVV
TH_0106|M.thermoresistible__bu GHGVGLHIHEAPGINAAAVGTLLAGSVVTVEPGVYLPDRGGVRIEDTLVV
MMAR_2180|M.marinum_M GHGVGLQIHEAPGIGATSVGTLLAGSVVTVEPGVYLPGRGGVRIEDTLVV
MUL_1764|M.ulcerans_Agy99 GHGVGLQIHEAPGIGATSVGTLLAGSVVTVEPGVYLPGRGGVRIEDTLVV
Mb2564c|M.bovis_AF2122/97 GHGVGLQIHEAPGIGVTSAGTLLAGSVVTVEPGVYLPGRGGVRIEDTLVV
Rv2535c|M.tuberculosis_H37Rv GHGVGLQIHEAPGIGVTSAGTLLAGSVVTVEPGVYLPGRGGVRIEDTLVV
MLBr_00521|M.leprae_Br4923 GHGVGLEIHEAPGIGTTSAGTLLAGSVVTVEPGVYLSGRGGVRIEDTLVV
MAV_3412|M.avium_104 GHGVGLEIHEAPGIGATSTGTLRAESVVTVEPGVYLPGRGGVRIEDTLVV
MAB_2838c|M.abscessus_ATCC_199 GHGVGLQIHEAPGIGSTATGTLLAGSAVTVEPGVYLPDRGGVRIEDTLVV
******.******:. ::.*** * *.*********..************
Mflv_3753|M.gilvum_PYR-GCK ------------------DKHPELLTRFPKELVII-
Mvan_2650|M.vanbaalenii_PYR-1 ------------------NKRPELLTRFPKELVII-
MSMEG_3034|M.smegmatis_MC2_155 PGGESAEGVVDRFAHNISDAAPELLTRFPKELAIV-
TH_0106|M.thermoresistible__bu G-----------------ETTPDLLTRFPKDLAVLD
MMAR_2180|M.marinum_M PN-GGPKTPRSAAQT------PELLTRFPKELTIL-
MUL_1764|M.ulcerans_Agy99 PN-GGPKTPRSAAQT------PELLTRFPKELTIL-
Mb2564c|M.bovis_AF2122/97 AG-GTPKMPETAGQT------PELLTRFPKELAIL-
Rv2535c|M.tuberculosis_H37Rv AG-GTPKMPETAGQT------PELLTRFPKELAIL-
MLBr_00521|M.leprae_Br4923 AAKGTSETPITSRPTG---FIPELLTRFPKELAIL-
MAV_3412|M.avium_104 PP-------ETAGKP------PELLTRFPKELAVLA
MAB_2838c|M.abscessus_ATCC_199 RP-----------------DGPELLTRFPKELIVID
*:*******:* ::