For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTELDLFVSPLGRVEGDLDVRVTINDGVVTSAWTEAAMFRGFEIILRGKDPQAGLIVCPRICGICGGSHL YKSAYALDTAWRTHMPPNATLIRNICQACETLQSIPRYFYALFAIDLTNKNYAKSKLYDEAVRRFAPYVG TSYQPGVVLSAKPVEVYAIFGGQWPHSSFMVPGGVMSAPTLSDVTRAIAILEHWNDNWLEKQWLGCSVDR WLENKTWNDVLAWVDENESQYNSDCGFFIRYCLDVGLDKYGQGVGNYLATGTYFEPSLYENPTIEGRNAA LIGRSGVFADGRYFEFDQANVTEDVTHSFYEGNRPLHPFEGETIPVNPEDGRRQGKYSWAKSPRYAVPGL GNVPLETGPLARRMAASAPDAETHQDDDPLFADIYNAIGPSVMVRQLARMHEGPKYYKWVRQWLDDLELK ESFYTKPVEYAEGKGFGSTEAARGALSDWIVIEDSKIKNYQVVTPTAWNIGPRDASEVLGPIEQALVGSP IVDAEDPVELGHVARSFDSCLVCTVHAYDGKTGKELNRFVINGMV
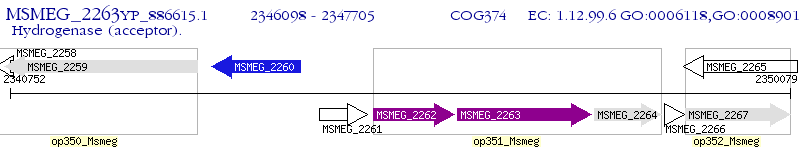
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2263 | hybC | - | 100% (535) | hydrogenase-2, large subunit |
| M. smegmatis MC2 155 | MSMEG_2719 | - | 4e-32 | 24.59% (606) | hydrogen:quinone oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_3928 | - | 7e-07 | 25.13% (199) | [NiFe] hydrogenase, alpha subunit, putative [Mycobacterium |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1869 | hyaB | 5e-35 | 24.88% (607) | nickel/iron-hydrogenase I large subunit, HyaB |
| M. avium 104 | MAV_2682 | - | 6e-31 | 25.42% (590) | hydrogen:quinone oxidoreductase |
| M. thermoresistible (build 8) | TH_1030 | hybC | 0.0 | 88.22% (535) | hydrogenase-2, large subunit |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2366 | - | 0.0 | 91.57% (534) | hydrogen:quinone oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2263|M.smegmatis_MC2_155 ---------------MTELDLFVSPLGRVEGDLDVR--VTINDGVVTSAW
Mvan_2366|M.vanbaalenii_PYR-1 --------------MASKLDLFVSPLGRVEGDLDVR--VTIEDGVVTSAW
TH_1030|M.thermoresistible__bu ---------------MTSLDLFVSPLGRVEGDLDVR--VTIEDGVVTSAW
MMAR_1869|M.marinum_M MTTIVPEPTTAKREPGQLVEMAWDPITRIVGSLGIYTKIDFENREVVECH
MAV_2682|M.avium_104 MTTIIPKPSHAKRGPGQLVEMAWDPITRIVGSLGIYTKIDFDNREVVECH
::: .*: *: *.*.: : ::: *...
MSMEG_2263|M.smegmatis_MC2_155 TEAAMFRGFEIILRGKDPQAGLIVCPRICGICGGSHLYKSAYALDTAWRT
Mvan_2366|M.vanbaalenii_PYR-1 TEAAMFRGFEIILQGKDPQSGLVVCPRICGICGGSHLYKSAYALDTAWRT
TH_1030|M.thermoresistible__bu TEAAMFRGFEIILRGKDPQAGLVVCPRICGICGGSHLYKSAYALDTAWRT
MMAR_1869|M.marinum_M STSSIFRGYSIFMKGKDPRDAHFITSRICGICGDNHATCSCYAQNMAYGV
MAV_2682|M.avium_104 STSSIFRGYSIFMKGKDPRDAHFITSRICGICGDNHCTCSVYTQNMAYGV
: :::***:.*:::****: . .: .*******..* * *: : *: .
MSMEG_2263|M.smegmatis_MC2_155 HMPPNATLIRNICQACETLQSIPRYFYALFAIDLTNKNYAKS--------
Mvan_2366|M.vanbaalenii_PYR-1 HMPHNATLVRNICQACETLQSIPRYFYALFAIDLTNKNYAKS--------
TH_1030|M.thermoresistible__bu HMPPNATLIRNICQACETLQSIPRYFYALFAIDLTNKNYAKS--------
MMAR_1869|M.marinum_M RPPHLGEWIVNLGEAAEYMFDHNIFQENLVGVDFCEKMVSETNPGVLAKA
MAV_2682|M.avium_104 QPPHLAEWITNLGEAAEYMFDHNIFQENLVGVDYCEKMVKETNPGVLAKA
: * . : *: :*.* : . : *..:* :* ::
MSMEG_2263|M.smegmatis_MC2_155 -------------KLYDEAVRRFAPYVGTSYQPGVVLSAKPVEVYAIFGG
Mvan_2366|M.vanbaalenii_PYR-1 -------------ALYDEAVRRFAPYVGTSYQPGVVLSAKPVEVYAIYGG
TH_1030|M.thermoresistible__bu -------------PLYDEAVRRFAPYVGTSYQTGVVLSAKPVEVYAIFGG
MMAR_1869|M.marinum_M EKTEAPHADAHGYKTIADIMRSLNPFSGEFYREALQVSRWTREMFCLMEG
MAV_2682|M.avium_104 ENTAAPHAGAHGYKTIADIMRSLNPFSGEFYREALQVSRMTREMFCLMEG
: :* : *: * *: .: :* . *::.: *
MSMEG_2263|M.smegmatis_MC2_155 QWPHSSFMVPGGVMSAPTLSDVTRAIAILEHWNDNWLEKQWLG-CSVDRW
Mvan_2366|M.vanbaalenii_PYR-1 QWPHSSFMVPGGVMCAPTLSDVTRSIAILEHWNDAWLEGQWLG-CSVDRW
TH_1030|M.thermoresistible__bu QWPHSSFMVPGGVMSAPTLADVTRSIAILEHWKDNWLEKQWLG-CSVERW
MMAR_1869|M.marinum_M RHVHPSTLYPGGVGTVATIQLMTDYMTRLMRYVEFMKKVVPLHDDLFDFF
MAV_2682|M.avium_104 RHVHPSTLYPGGVGTVATIQLMTDYMSRLMRYVEFMKKVVPMHDDLFDFF
: *.* : **** ..*: :* :: * :: : : : .: :
MSMEG_2263|M.smegmatis_MC2_155 LENKTWNDVLAWVDENESQYNSDCGFFIRYCLDVGLDKYGQGVGNYLATG
Mvan_2366|M.vanbaalenii_PYR-1 LENKTWNDVLAWVDENEAQYNSDCGFFIRYCLDVGLDKYGQGVGNYLATG
TH_1030|M.thermoresistible__bu LENKTWDDVLAWVDENEAQYNSDCGFFIRYCLDIGLDKYGQGVGNYLATG
MMAR_1869|M.marinum_M YDALPGYEKVGLRRTLLGCWGSFQDPEVCNFEYKDMERWGN--AMFVTPG
MAV_2682|M.avium_104 YEALPGYEKSGLRRTLLACWGSYQDPEHCNFSYKDMEKWGR--KMFVTPG
: . : . . :.* . .::::*. :::.*
MSMEG_2263|M.smegmatis_MC2_155 TYFEPSLYENPTIEGRNAALIGRSGVFADGRYFEFDQANVTEDVTHSFYE
Mvan_2366|M.vanbaalenii_PYR-1 TYFEPSLYENPTIDGRNAALIGRSGVYADGRYFEFDQANVLEDVTHSFYQ
TH_1030|M.thermoresistible__bu TYFEPSRYENPTIEGRNDALIGRSGIFADGKWYEFDQSRVTEDVTHSFFQ
MMAR_1869|M.marinum_M VVVDGKLVTHSLVDIN----LGIRILLGSSYYDDWTDQEMFVKTDPLGNK
MAV_2682|M.avium_104 IVVDGKLVTTSLVDIN----LGIRILLGSSYYDDWTDQEMFVKTDPLGNP
.: . . :: . :* : ... : :: : .: ..
MSMEG_2263|M.smegmatis_MC2_155 GNRPLHPFEGETIPVNPEDGRRQGKYSWAKSPRYAVPGLGNVPLETG--P
Mvan_2366|M.vanbaalenii_PYR-1 GDRPLHPFDGETIPIDPEQGRKQGKYSWAKSPRYAVGDLGNIPLEAG--P
TH_1030|M.thermoresistible__bu GDRPLHPFEGETIPIDPEVGRAQGKYSWAKSPRYDIPEVGRIPLEAG--P
MMAR_1869|M.marinum_M VDR-RHPWNQHTNPHPQKREMDGGKYSWVMSPRWFD-GKDHLALDTGGGP
MAV_2682|M.avium_104 VDR-RHPWNQHTNPHPQKRDMED-KYSWVMSPRWFD-GKDHLALDTGGGP
:* **:: .* * : ****. ***: ..:.*::* *
MSMEG_2263|M.smegmatis_MC2_155 LARRMAASAP------DAETHQDDDPLFADIYNAIGPSVMVRQLARMHEG
Mvan_2366|M.vanbaalenii_PYR-1 LARRMAAAGP------DAAAHQDNDPLFGDIYNTIGPSVMVRQLARMHEA
TH_1030|M.thermoresistible__bu LARRMAAGGP------NALEHQDNDPLFVDIYNKIGPSVMVRQLARMHEA
MMAR_1869|M.marinum_M LARLWATALAGLVDIGYVKATGNSVKINLPKTALKGPVEFEWKVPKHGSN
MAV_2682|M.avium_104 LARLWSTALAGLVDIGYVKATGRSVQINLPKTALKGPVELEWKIPEHGSN
*** ::. . . . : ** : ::.. .
MSMEG_2263|M.smegmatis_MC2_155 PKYYKWVRQWLDDLELKESFYTKPVEYAE-----------------GKGF
Mvan_2366|M.vanbaalenii_PYR-1 PKYYKWTRQWLDALELKESFYTKPTEYAE-----------------GKGF
TH_1030|M.thermoresistible__bu PKYYQWTRAWLDQLDLKESFYTKPVEHAE-----------------GRGF
MMAR_1869|M.marinum_M TIERDRARTYFQAYAAACALHFAEKALAEIRAGRTKTWEKFEVPDEGIGC
MAV_2682|M.avium_104 TIERNRARTYFQAYSAAAALHFAEKALVEIRAGRTKTWEKFEVPDEGIGC
. . .* ::: ::: .* * *
MSMEG_2263|M.smegmatis_MC2_155 GSTEAARGALSDWIVIEDSKIKNYQVVTPTAWNIGPRDASEVLGPIEQAL
Mvan_2366|M.vanbaalenii_PYR-1 GSTEAARGALSDWIVIEDSKIKNYQVVTPTAWNIGPRDGSEVLGPIERAL
TH_1030|M.thermoresistible__bu GATEAARGALADWIVIEDSKIANYQVITPTAWNIGPRDASETLGPIEKAL
MMAR_1869|M.marinum_M GFTEAVRGVLSHHLVIRNGKIANYHPYPPTPWNANPRDSFGTPGPYEDAV
MAV_2682|M.avium_104 GFTEAVRGVLSHHMVIRDGKIANYHPYPPTPWNANPRDVYGTPGPYEDAV
* ***.**.*:. :**.:.** **: .**.** .*** . ** * *:
MSMEG_2263|M.smegmatis_MC2_155 VGSPIVDAEDP-----VELGHVARSFDSCLVCTVHAYDGKTGKELNRFVI
Mvan_2366|M.vanbaalenii_PYR-1 VGSPIVDAEDP-----VELGHVARSFDSCLVCTVHAYDGKTGRELSKFVI
TH_1030|M.thermoresistible__bu VGAPVVDAEDP-----VELGHVARSFDSCLVCTVHAYDGRSGRELSKFVI
MMAR_1869|M.marinum_M QGQPIFEENDRENFKGIDVMRTVRSFDPCLPCGVHMYLGK-GKTLERLHT
MAV_2682|M.avium_104 QGQPIFEENDRDNFKGIDIMRTVRSFDPCLPCGVHMYLGK-GKTLDRLHS
* *:.: :* ::: :..****.** * ** * *: *: *.::
MSMEG_2263|M.smegmatis_MC2_155 NGMV----
Mvan_2366|M.vanbaalenii_PYR-1 NGMV----
TH_1030|M.thermoresistible__bu NGMV----
MMAR_1869|M.marinum_M PTQSPVGE
MAV_2682|M.avium_104 PTQTATGE