For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTESPAGRAIDCLANVHFGETQNQPAFMTKVRDDYFKGPKSMYDPIDLGALLEEMDANGVERAILMDNLT KPSVTARKFVEAHPERFALAMGGVNLLRPIPSLRELAAAVRDLPVAYAVVGPSFWGDGQYPPSDAVYYPL YTKCAELDLPLCVNTGLPGPPIPGEVQNPIHLDRVCVRFPELRLCMIHGADPWWDIAIRMLIKYKNLRLM TSAWSPKRLPESLLHYMRTRGPDKIIFASDFPVLRMNRVVPEARALDLPPDVLDNYLYHNAAEFFFGGRQ PQEF
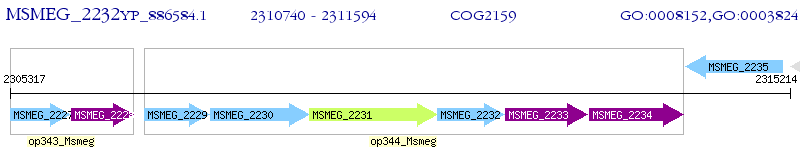
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2232 | - | - | 100% (284) | amidohydrolase family protein |
| M. smegmatis MC2 155 | MSMEG_0628 | - | 4e-06 | 25.95% (158) | amidohydrolase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3540c | - | 5e-28 | 32.08% (240) | hypothetical protein Mb3540c |
| M. gilvum PYR-GCK | Mflv_4348 | - | 1e-137 | 82.22% (270) | amidohydrolase 2 |
| M. tuberculosis H37Rv | Rv3510c | - | 5e-28 | 32.08% (240) | hypothetical protein Rv3510c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4667 | - | 1e-07 | 27.39% (157) | putative amidohydrolase |
| M. marinum M | MMAR_4012 | - | 1e-132 | 80.00% (270) | metal-dependent hydrolase |
| M. avium 104 | MAV_1593 | - | 1e-139 | 83.15% (273) | amidohydrolase family protein |
| M. thermoresistible (build 8) | TH_1218 | - | 1e-140 | 82.91% (275) | amidohydrolase family protein |
| M. ulcerans Agy99 | MUL_3876 | - | 1e-130 | 78.89% (270) | metal-dependent hydrolase |
| M. vanbaalenii PYR-1 | Mvan_1996 | - | 1e-136 | 80.36% (275) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3540c|M.bovis_AF2122/97 ---------------MTIDVWMQHPTQRFLHGDMFASLRR---WTGGSIP
Rv3510c|M.tuberculosis_H37Rv ---------------MTIDVWMQHPTQRFLHGDMFASLRR---WTGGSIP
MMAR_4012|M.marinum_M --------MTD----RAIDCLVNVHFGEAEAQPNWMLKVRDDYFKGPDSM
MUL_3876|M.ulcerans_Agy99 --------MTD----RAIDCLVNVHFGEAEAQPNWMLKVRDDYFKGPDSM
MAV_1593|M.avium_104 ----------M----KAIDCLVNVHFGETEKQPTWMLKVRDDYFKGPASM
TH_1218|M.thermoresistible__bu --------MTR----KVLDCLVNVHFGEAEMQPAWMEKVRDDYFKGPEAM
Mflv_4348|M.gilvum_PYR-GCK --------MSD----RVIDCLVNVHFGETANQPEFMLKVRDDYFKGPQSL
Mvan_1996|M.vanbaalenii_PYR-1 --------MTH----KVIDCLVNVHFGETAKQPEFMLKVRDDYFKGPQSL
MSMEG_2232|M.smegmatis_MC2_155 --------MTESPAGRAIDCLANVHFGETQNQPAFMTKVRDDYFKGPKSM
MAB_4667|M.abscessus_ATCC_1997 MSESAFDPAPVRQIWQELGIPGIVDVHTHFMPKSVMDKVWAYFDSAGPLV
:. ..
Mb3540c|M.bovis_AF2122/97 ETDIPIEATVSSMDAGGVTLGLLSAWRGPNGQDLISNDAVAEWVRLYPNR
Rv3510c|M.tuberculosis_H37Rv ETDIPIEATVSSMDAGGVTLGLLSAWRGPNGQDLISNDAVAEWVRLYPNR
MMAR_4012|M.marinum_M FAPVDLAQLLDEMDEQGVQRAILMDSLAS------PSITARKFVQAKPDR
MUL_3876|M.ulcerans_Agy99 FAPVDLAQLLDEMDEQGVQRAILMDSLAS------PSLTACKFVQAKPDR
MAV_1593|M.avium_104 FAPVELSELIDEMDAQGVRKAILMDNLAK------PSVTARKFVEDKPER
TH_1218|M.thermoresistible__bu FAPVDLAELLDEMDEQGVQKAILMDNLSK------PSVTARKFVEARPDR
Mflv_4348|M.gilvum_PYR-GCK YDQVELPALLDEMAEHGVEKAILMDNLVK------PSVTARKFAEERPDK
Mvan_1996|M.vanbaalenii_PYR-1 YEQVELPALLDEMAEHGVEKAILMDNLVK------PSVTARKFVEERPDK
MSMEG_2232|M.smegmatis_MC2_155 YDPIDLGALLEEMDANGVERAILMDNLTK------PSVTARKFVEAHPER
MAB_4667|M.abscessus_ATCC_1997 GRPWPITYRADESQRVTTLRDFGVLRFTSLVYP--HKPQMAAWLNQWAAQ
: .. . : . : . . :
Mb3540c|M.bovis_AF2122/97 FAG-LAAVDLDRPMAAVRELRRRVGEGFVGLRVV-PWLWG---APPTDRR
Rv3510c|M.tuberculosis_H37Rv FAG-LAAVDLDRPMAAVRELRRRVGEGFVGLRVV-PWLWG---APPTDRR
MMAR_4012|M.marinum_M FALAMGGINLLRPVRPLRELTAIVGELPVAYAAVGPSFWGEGQYPPSDAV
MUL_3876|M.ulcerans_Agy99 FALAMGGINLLRPVRPLRELTAIVGELPVAYAAVGPSFWGEGQYPPSDAV
MAV_1593|M.avium_104 FALAMGGVNLLRPIPSLRELGAVVADLPVAYAAVGPSFWGDGQYPPSDAV
TH_1218|M.thermoresistible__bu FALAMGGVNLLRPIPSLRELAAVAADLPVVYAVVGPSFWGDGQYPPSDAV
Mflv_4348|M.gilvum_PYR-GCK FALAIGGVNLLRPMPSLRELSSVVADLPVAYAAVGPSFWGDGMYPPSDAV
Mvan_1996|M.vanbaalenii_PYR-1 FALAVGGVNLLRPMPSLRELAAVVADLPVAYAAVGPSFWGDGMYPPSDAV
MSMEG_2232|M.smegmatis_MC2_155 FALAMGGVNLLRPIPSLRELAAAVRDLPVAYAVVGPSFWGDGQYPPSDAV
MAB_4667|M.abscessus_ATCC_1997 FAAQTPDCIHTATFFPEPGAADYVREAIDAGVGVFKVHVQVGAFSPTDPL
** .. . . : * .*:*
Mb3540c|M.bovis_AF2122/97 YYPLFAECVQSAVPFCTQVGHTGPLRPSETGRPIPYIDQVALDFPELVIV
Rv3510c|M.tuberculosis_H37Rv YYPLFAECVQSAVPFCTQVGHTGPLRPSETGRPIPYIDQVALDFPELVIV
MMAR_4012|M.marinum_M YYPLYAKCAELELALCINTGIPGPPIPGEVQHPM-HLDRVCVRFPELRLC
MUL_3876|M.ulcerans_Agy99 YYPLYVKCAELELALCINTGIPGPPIPGEVQHPM-HLDRVCVRFPELRLC
MAV_1593|M.avium_104 YYPLYTKCAELELPLCVNTGIPGPPIPGEVQNPI-HLDRVCVRFPELKLC
TH_1218|M.thermoresistible__bu YYPLYTKCAELELPLCVNTGIPGPPIPGEVQNPI-HLDRVCVRFPELRLC
Mflv_4348|M.gilvum_PYR-GCK YYPLYTKCAELGLPLCINTGLPGPPIPGEVQNPI-HLDRVCVRFPELKLC
Mvan_1996|M.vanbaalenii_PYR-1 YYPLYTKCAELELPLCINTGLPGPPIPGEVQNPI-HLDRVCVRFPELKLC
MSMEG_2232|M.smegmatis_MC2_155 YYPLYTKCAELDLPLCVNTGLPGPPIPGEVQNPI-HLDRVCVRFPELRLC
MAB_4667|M.abscessus_ATCC_1997 LVPVWGLIEDAGVPVVIHCGS-GP-APGEFTGPD-PVTALLKQFPHLRLI
*:: : :.. : * ** *.* * : : **.* :
Mb3540c|M.bovis_AF2122/97 CGHVGYPWTEEMVAVARKHENVYIDT----SAYTIKRLPGKLVRFMKTDT
Rv3510c|M.tuberculosis_H37Rv CGHVGYPWTEEMVAVARKHENVYIDT----SAYTIKRLPGKLVRFMKTDT
MMAR_4012|M.marinum_M MIHGADPWWDVAIRLLLKYSNLRLMT----SAWSPKRLPESLLHYMRTR-
MUL_3876|M.ulcerans_Agy99 MIHGADPWWDVAIRLLLKYSNSRLMT----SAWTPKRLPESLLHYMRTR-
MAV_1593|M.avium_104 MIHGADPWWDVAIRLLLKYANLRLMT----SAWSPKRLPESLLHYMRTR-
TH_1218|M.thermoresistible__bu MIHGADPWWDIAIRLLIKYQNLRLMT----SAWSPKRLPNSLLHYMRTR-
Mflv_4348|M.gilvum_PYR-GCK MIHGADPWWDVAIRLLIKYANLRIMT----SAWSPKRLPDSLLHYMRTR-
Mvan_1996|M.vanbaalenii_PYR-1 MIHGADPWWDVAIRLLIKYANLRIMT----SAWSPKRLPDSLLHFMRTR-
MSMEG_2232|M.smegmatis_MC2_155 MIHGADPWWDIAIRMLIKYKNLRLMT----SAWSPKRLPESLLHYMRTR-
MAB_4667|M.abscessus_ATCC_1997 VAHMGMPEYSEFLDLCEQHDEVRLDTTMAFTAFSNEKAPFPESDYPRLVE
* . * . : : :: : : * :*:: :: * : :
Mb3540c|M.bovis_AF2122/97 GQRKVLFGTNYPMIAH--THALTGLDELG-LSDEARRDFLHGNAVRVFKL
Rv3510c|M.tuberculosis_H37Rv GQRKVLFGTNYPMIAH--THALTGLDELG-LSDEARRDFLHGNAVRVFKL
MMAR_4012|M.marinum_M GKNKVIFASDWPVLRM--RRVVPEARALD-LPADVLANYLYNNAQEFFFG
MUL_3876|M.ulcerans_Agy99 GKNKVIFASDWPVLRM--RRVVPEARALD-LPADVLANYLYNNAQEFFFG
MAV_1593|M.avium_104 GQDKVIFASDWPVLKM--HRVVPEAAGLD-LPADVLENYLYNNAHEFFFG
TH_1218|M.thermoresistible__bu GPNKVIFGSDWPVLTM--RRVVPEAYALD-LPSEVLDNYLYNNAAEFFFG
Mflv_4348|M.gilvum_PYR-GCK GKNKVIFASDWPVLRQ--SRVVPEALALD-LPPDVLDNYLYNNANDFFFG
Mvan_1996|M.vanbaalenii_PYR-1 GKNKVIFASDWPVLRQ--SRVVPEAVALD-LPPDVLDNYLYNNANDFFFG
MSMEG_2232|M.smegmatis_MC2_155 GPDKIIFASDFPVLRM--NRVVPEARALD-LPPDVLDNYLYHNAAEFFFG
MAB_4667|M.abscessus_ATCC_1997 MGHKIYFGSDFPNIPYGYAEAITALRRMPGIDDAWMRDVLYHNGARLFGV
*: *.:::* : ..:. : : : *: *. .*
Mb3540c|M.bovis_AF2122/97 DPRGKVQT
Rv3510c|M.tuberculosis_H37Rv DPRGKVQT
MMAR_4012|M.marinum_M E-ESRR--
MUL_3876|M.ulcerans_Agy99 E-ESGR--
MAV_1593|M.avium_104 G-HKEG--
TH_1218|M.thermoresistible__bu GPEQER--
Mflv_4348|M.gilvum_PYR-GCK TDEQEI--
Mvan_1996|M.vanbaalenii_PYR-1 AAEQEI--
MSMEG_2232|M.smegmatis_MC2_155 GRQPQEF-
MAB_4667|M.abscessus_ATCC_1997 DTSASGR-