For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNARYTTDTLRTARSFLFVPGNRPERFSKAVAAQPDVVVLDLE
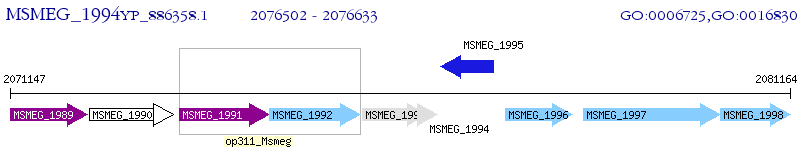
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1994 | - | - | 100% (43) | HpcH/HpaI aldolase |
| M. smegmatis MC2 155 | MSMEG_4713 | - | 8e-05 | 62.50% (32) | HpcH/HpaI aldolase/citrate lyase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2556 | - | 4e-05 | 57.14% (35) | HpcH/HpaI aldolase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_3845 | citE | 4e-05 | 65.71% (35) | citrate (pro-3s)-lyase (beta subunit) CitE |
| M. avium 104 | MAV_5074 | - | 2e-07 | 55.56% (36) | citrate lyase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3776 | citE | 2e-05 | 65.71% (35) | citrate (pro-3s)-lyase (beta subunit) CitE |
| M. vanbaalenii PYR-1 | Mvan_4087 | - | 3e-05 | 60.00% (35) | HpcH/HpaI aldolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2556|M.gilvum_PYR-GCK MTL-------APGTAWLFCPADRPERFEKAAAAA-DIVILDLEDGVAAKD
Mvan_4087|M.vanbaalenii_PYR-1 MTL-------AAGPAWLFCPADRPERFEKAAAAA-DVVILDLEDGVAAKD
MMAR_3845|M.marinum_M MKLR------TAGPAWLFCPADRPERFAKAAAAA-DVVILDLEDGVAEAQ
MUL_3776|M.ulcerans_Agy99 MKLR------TASPAWLFCPADRPERFAKAAAAA-DVVILDLEDGVAEAQ
MSMEG_1994|M.smegmatis_MC2_155 MNARYTTDTLRTARSFLFVPGNRPERFSKAVAAQPDVVVLDLE-------
MAV_5074|M.avium_104 MHDD-----LASARSLLFVPGDRPDRFTKAASSDADGIIIDLEDAVAPEA
* .. : ** *.:**:** **.:: * :::***
Mflv_2556|M.gilvum_PYR-GCK RQAAREALVNTPLDPARTVVRVNPTGTADHGPDLEAVARTAYTTVMLAKT
Mvan_4087|M.vanbaalenii_PYR-1 REAARKALVDTPLDPSRTVVRVNPAGTPDHKLDLEALSRTSYTTVMLAKT
MMAR_3845|M.marinum_M KPAARQALLDTPLDPERTIVRINAGDTDEQARDLAALAGTAYTTVMLPKA
MUL_3776|M.ulcerans_Agy99 KPAARQALLDTPLDPERTIVRINAGDTDEQARDLAALAGTAYTTVMLPKA
MSMEG_1994|M.smegmatis_MC2_155 --------------------------------------------------
MAV_5074|M.avium_104 KARAREAVDRWLATGGVGTVRINGTETPWHQEDLEMAARHG-CPVMVPKA
Mflv_2556|M.gilvum_PYR-GCK EAPEQVRALAP-----LDVIVLIETPLGALNVVDLARVDNAYALMWGAED
Mvan_4087|M.vanbaalenii_PYR-1 ESPQQVAALAP-----LDVVVLIETPLGALAVVELARVDNALALMWGAED
MMAR_3845|M.marinum_M ESAAQLALLAP-----RDVIALVETARGALFAAEIAAADNTVGMMWGAED
MUL_3776|M.ulcerans_Agy99 ESVAQLALLAP-----RDVIALVETARGALFAAEIAAADNTVGMMWGAED
MSMEG_1994|M.smegmatis_MC2_155 --------------------------------------------------
MAV_5074|M.avium_104 RSASHLAQISRRLPTGTALIALIETAAGVLAAADICAVEGVVRAAFGSVD
Mflv_2556|M.gilvum_PYR-GCK LFAELGGTANRYPDGSYREVARHVRSQTLLAAKAYGRVA-LDSVFLDIKN
Mvan_4087|M.vanbaalenii_PYR-1 LFAVLGGTANRYPDGSYREVARHVRSQTLLAAKAYGRLA-LDSVFLDIKN
MMAR_3845|M.marinum_M LIATLGGSSSRRGDGSYRDVARHVRSTTLLAASAFGRLA-LDAVHLDIGD
MUL_3776|M.ulcerans_Agy99 LIATLGGSSSRRGDGTYRDVARHVRSTILLAASAFGRLA-LDAVHLDIGD
MSMEG_1994|M.smegmatis_MC2_155 --------------------------------------------------
MAV_5074|M.avium_104 LGAELG------IDPEDHESLRYARSAVVLGSAAAGLAAPLDGVTTAVHD
Mflv_2556|M.gilvum_PYR-GCK LDGLRGEVDDAVAVGFDGKVAIHPTQLPVIRDGYTPTEAQADWARRVLAA
Mvan_4087|M.vanbaalenii_PYR-1 LDGLRAEVDDAVAVGFDVKVAIHPSQVAVIRSGYAPTDAEVDWARRVLDT
MMAR_3845|M.marinum_M IDGLHAEAIDAATVGFDVTVCIHPSQVEVVRKAYRPDEEKLAWARRVLVA
MUL_3776|M.ulcerans_Agy99 VDGLHAEAMDAATVGFDVTVCIHPSQVEVVRKAYRPDEEKLAWARRVLVA
MSMEG_1994|M.smegmatis_MC2_155 --------------------------------------------------
MAV_5074|M.avium_104 EGAAGADAARAARLGFGGKLCIHPCQVAPVNSQFSPSIEEQAWARKVMAA
Mflv_2556|M.gilvum_PYR-GCK AQAERGVFQYEGQMVDMPVLRRAERIVALAP-----
Mvan_4087|M.vanbaalenii_PYR-1 ARTQRGVFAFEGIMVDAPVLRRAERIVELAPHPGA-
MMAR_3845|M.marinum_M ARTERGVFAFEGQMVDSPVLRHAEMLLRRAGESVPA
MUL_3776|M.ulcerans_Agy99 ARTERGVFAFEGQMVDSPVLRHAEMLLRRAGESVPA
MSMEG_1994|M.smegmatis_MC2_155 ------------------------------------
MAV_5074|M.avium_104 G-SGNGARTVDGCMVDKPVVDRAARILARAQGLTSP