For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVNRYKVAVIAGDGIGREVVPEGLRVLQAASERFGFTLDLEEFDYANVDYYLKHGQMMPDDWFEQLRGFD AIFFGAVGWPELVPDHISLWGSLIQFRRHFDQYVNLRPVRLLPGIAGPLAGRKPGDIDFYVVRENTEGEY SSIGGKIFEDTPRETVIQDTVMTRVGVDRILRYAFELAQRRPRRLLTSATKSNGISISMPYWDERVEQMS TMFPDVSVNKFHIDILAANFVLHPDWFDVVVASNLFGDILSDLGPACTGTIGVAPSANINPERTFPSLFE PVHGSAPDIAGQGVANPVGQIWCASMMLEHLGERTAAKAVLDAIEAVLADGAVLTPDLGGTATTTALGTA IADSVGASASASAGTR
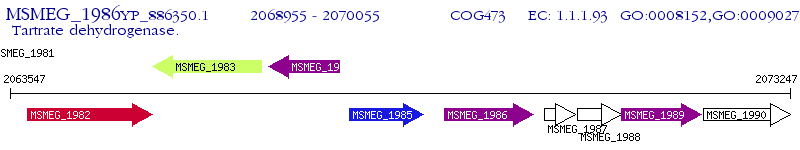
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1986 | - | - | 100% (366) | tartrate dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2379 | leuB | 1e-68 | 44.75% (324) | 3-isopropylmalate dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3285 | - | 2e-42 | 34.08% (355) | 3-isopropylmalate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3019c | leuB | 3e-67 | 44.34% (327) | 3-isopropylmalate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4233 | - | 3e-65 | 43.83% (324) | 3-isopropylmalate dehydrogenase |
| M. tuberculosis H37Rv | Rv2995c | leuB | 5e-67 | 44.34% (327) | 3-isopropylmalate dehydrogenase |
| M. leprae Br4923 | MLBr_01691 | leuB | 4e-63 | 41.84% (337) | 3-isopropylmalate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4419 | - | 2e-99 | 55.40% (352) | tartrate dehydrogenase |
| M. marinum M | MMAR_1716 | leuB | 8e-65 | 43.77% (329) | 3-isopropylmalate dehydrogenase LeuB |
| M. avium 104 | MAV_3845 | leuB | 6e-68 | 44.67% (347) | 3-isopropylmalate dehydrogenase |
| M. thermoresistible (build 8) | TH_2038 | leuB | 3e-64 | 41.54% (337) | PROBABLE 3-ISOPROPYLMALATE DEHYDROGENASE LEUB (BETA-IPM |
| M. ulcerans Agy99 | MUL_1953 | leuB | 3e-64 | 43.16% (329) | 3-isopropylmalate dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_2129 | - | 7e-66 | 44.14% (324) | 3-isopropylmalate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01691|M.leprae_Br4923 ----MKLAIIGGDGIGPEVVAQAVKILDVVLPGV----QKTTYDLG-ARR
MAV_3845|M.avium_104 ----MKLAVIGGDGIGPEVTAEALKVLDAVLPGV----DKTEYDLG-ARR
Mb3019c|M.bovis_AF2122/97 ----MKLAIIAGDGIGPEVTAEAVKVLDAVVPGV----QKTSYDLG-ARR
Rv2995c|M.tuberculosis_H37Rv ----MKLAIIAGDGIGPEVTAEAVKVLDAVVPGV----QKTSYDLG-ARR
MMAR_1716|M.marinum_M ----MKLAIVAGDGIGPEVVAQAVKVLDVVQPGV----EKTNYDLG-ARR
MUL_1953|M.ulcerans_Agy99 ----MKLAIVAGDGIGPEVVAQAVKILDVVQPGV----EKTNYDLG-ARR
TH_2038|M.thermoresistible__bu --VTMKLAIIPGDGIGPEVIGEAVKVLDAVLPDV----DKTHFDLG-ARR
Mflv_4233|M.gilvum_PYR-GCK ----MKLAVIAGDGIGPEVIGEALRVLDAVVPGV----EKTEYDLG-ARL
Mvan_2129|M.vanbaalenii_PYR-1 ----MRLAVIAGDGIGPEVITEALKVLDAVLPGV----EKTEYDLG-ARQ
MSMEG_1986|M.smegmatis_MC2_155 MVNRYKVAVIAGDGIGREVVPEGLRVLQAASERFGFTLDLEEFDYANVDY
MAB_4419|M.abscessus_ATCC_1997 MSDRMRVATIPGDGIGVDVTREAVKVMDSATGGR---IEWTEFDYS-CER
::* : ***** :* :.::::: . : :* .
MLBr_01691|M.leprae_Br4923 YHTTGELLPESVLAELREHDAILLGAVGDPS-VPSGVLERGLLLRLRFEL
MAV_3845|M.avium_104 YHATGELLPDSVIDELRAHDAILLGAIGDPS-VPSGVLERGLLLRLRFEL
Mb3019c|M.bovis_AF2122/97 FHATGEVLPDSVVAELRNHDAILLGAIGDPS-VPSGVLERGLLLRLRFEL
Rv2995c|M.tuberculosis_H37Rv FHATGEVLPDSVVAELRNHDAILLGAIGDPS-VPSGVLERGLLLRLRFEL
MMAR_1716|M.marinum_M FHATGEILPDSVIAELREHDAILLGAIGDPS-VPSGVLERGLLLRLRFEL
MUL_1953|M.ulcerans_Agy99 FHATGEILPDSVIAELREHDAILLGAIGDPS-VPSGVLERGLLLRLRFEL
TH_2038|M.thermoresistible__bu FHATGEVLPDSVIDELRAHDAILLGAIGDPS-VPSGVLERGLLLRIRFEL
Mflv_4233|M.gilvum_PYR-GCK YHRTGEVLPDSVLDELKGHDAILLGAIGDPS-MPSGVLERGLLLRIRFEL
Mvan_2129|M.vanbaalenii_PYR-1 YHKTGEVLPDSVLEELKGHDAILLGAIGDPS-VPSGLLERGLLLRIRFEL
MSMEG_1986|M.smegmatis_MC2_155 YLKHGQMMPDDWFEQLRGFDAIFFGAVGWPELVPDHISLWGSLIQFRRHF
MAB_4419|M.abscessus_ATCC_1997 YAKTGAMMPPDAPTTLAGFDAILLGAVGYPG-VPDDVSLWGLLIPLRRAF
: * ::* . * .***::**:* * :*. : * *: :* :
MLBr_01691|M.leprae_Br4923 DHHINLRPARLYPGVNSLLA---GKPDIDFVVVREGTEGPYTGTGGAIRV
MAV_3845|M.avium_104 DHHINLRPGRLYPGVSSPLA---GNPDIDFVVVREGTEGPYTGTGGAIRV
Mb3019c|M.bovis_AF2122/97 DHHINLRPARLYPGVASPLS---GNPGIDFVVVREGTEGPYTGNGGAIRV
Rv2995c|M.tuberculosis_H37Rv DHHINLRPARLYPGVASPLS---GNPGIDFVVVREGTEGPYTGNGGAIRV
MMAR_1716|M.marinum_M DHHINLRPGRLYPGVKSPLALEPGNPEIDFVVVREGTEGPYTGNGGAIRV
MUL_1953|M.ulcerans_Agy99 DYHINLRPGRLYPGVKSPLALEPGNPEIDFVVVREGTEGPYTGNGGAIRV
TH_2038|M.thermoresistible__bu DHHINLRPARLYEGVSSPLA---GNPDIDFVVVREGTEGPYTGTGGAIRV
Mflv_4233|M.gilvum_PYR-GCK DHHINLRPGRLYPGVQSPLA---GNPEIDFVVVREGTEGPYTGNGGAIRV
Mvan_2129|M.vanbaalenii_PYR-1 DHHINLRPARLYPGVQSPLA---GNPEIDFVVVREGTEGPYTGNGGAIRV
MSMEG_1986|M.smegmatis_MC2_155 DQYVNLRPVRLLPGIAGPLAGR-KPGDIDFYVVRENTEGEYSSIGGKIFE
MAB_4419|M.abscessus_ATCC_1997 NQYVNLRPVRLLPGIASPLAGV-SDEALSMVIVRENTEGEYSPIGGVHNR
: ::**** ** *: . *: :.: :***.*** *: **
MLBr_01691|M.leprae_Br4923 GTPNEVATEVSVNTAFGVRRVVQDAFERARQ-RRKHLTLVHKNNVLTYAG
MAV_3845|M.avium_104 GTPNEVATEVSQNTAFGVRRVVVDAFERARR-RRKHLTLVHKTNVLTFAG
Mb3019c|M.bovis_AF2122/97 GTPNEVATEVSVNTAFGVRRVVADAFERARR-RRKHLTLVHKTNVLTLAG
Rv2995c|M.tuberculosis_H37Rv GTPNEVATEVSVNTAFGVRRVVADAFERARR-RRKHLTLVHKTNVLTFAG
MMAR_1716|M.marinum_M GTANEVATEVSVNTAFGVRRVVRDAFERAMR-RRKHLTLVHKNNVLTFAG
MUL_1953|M.ulcerans_Agy99 GTANEVATEVSVNTAFGVRRVVRDAFERAMR-RRKHLTLVHKNNVLTFAG
TH_2038|M.thermoresistible__bu GTPNEVATEVSLNTAFGVRRVVENAFQRAAQ-RRKHLTLVHKTNVLTYAG
Mflv_4233|M.gilvum_PYR-GCK GTPHEIATEVSVNTAYGVRRVVQDAFKRAQQ-RRKHLTLVHKNNVLTNAG
Mvan_2129|M.vanbaalenii_PYR-1 GTPHEIATEVSVNTAFGVRRAVQDAFGRAQQ-RRKHLTLVHKNNVLTNAG
MSMEG_1986|M.smegmatis_MC2_155 DTPRETVIQDTVMTRVGVDRILRYAFELAQRRPRRLLTSATKSNGISISM
MAB_4419|M.abscessus_ATCC_1997 GNQNEFALQESVFTRIGCERIIRYAFELAASAGFR-VCSATKSNGIIHSM
.. .* . : : * * * : ** * : : . *.* : :
MLBr_01691|M.leprae_Br4923 TLWCRIVQEVGEKYPDVEVVYQHIDAATIYLVTEPSRFDVIVTDNLFGDI
MAV_3845|M.avium_104 KLWSRIVAEVGRDYPDVEVAYQHIDAATIFMVTDPGRFDVIVTDNLFGDI
Mb3019c|M.bovis_AF2122/97 GLWLRTVDEVGECYPDVEVAYQHVDAATIHMITDPGRFDVIVTDNLFGDI
Rv2995c|M.tuberculosis_H37Rv GLWLRTVDEVGECYPDVEVAYQHVDAATIHMITDPGRFDVIVTDNLFGDI
MMAR_1716|M.marinum_M SLWWRTVQEIGEEYPDVELAYQHVDAATIHMVTDPGRFDVIVTDNLFGDI
MUL_1953|M.ulcerans_Agy99 SLWWRTVQEIGEEYPDVELAYQHVDAATIHMVTDPGRFDVIVTDNLFGDI
TH_2038|M.thermoresistible__bu ALWSRTVQEVGEQYPDVEVAYQHVDAATIHLVTDPGRFDVIVTDNLFGDI
Mflv_4233|M.gilvum_PYR-GCK SLWWRTVQAVAAEYPEVEVAYQHVDAATIHMVTDPGRFDVIVTDNLFGDI
Mvan_2129|M.vanbaalenii_PYR-1 SLWWRTVQTVAAEYPDVEVAYQHVDAATIHMVTDPGRFDVIVTDNLFGDI
MSMEG_1986|M.smegmatis_MC2_155 PYWDERVEQMSTMFPDVSVNKFHIDILAANFVLHPDWFDVVVASNLFGDI
MAB_4419|M.abscessus_ATCC_1997 PYWDSIFNEIAAEYPQTPSVSMHVDALAARMVSDPASLDVIVASNLFGDI
* . :. :*:. *:* : :: .* :**:*:.******
MLBr_01691|M.leprae_Br4923 ITDLAAAVCGGIALAASGNIDATRTNPSMFEPVHGSAPDIAGQGIADPTA
MAV_3845|M.avium_104 ITDLSAAVCGGIGLAASGNIDGTRTNPSMFEPVHGSAPDIAGQGVADPTA
Mb3019c|M.bovis_AF2122/97 ITDLAAAVCGGIGLAASGNIDATRANPSMFEPVHGSAPDIAGQGIADPTA
Rv2995c|M.tuberculosis_H37Rv ITDLAAAVCGGIGLAASGNIDATRANPSMFEPVHGSAPDIAGQGIADPTA
MMAR_1716|M.marinum_M ITDLAAAVCGGIGLAASGNIDATRTNPSMFEPVHGSAPDIAGQGIADPTA
MUL_1953|M.ulcerans_Agy99 ITDLAAAVCGGIGLAASGNIDATRTNPSMFEPVHGSAPDIAGQGIADPTA
TH_2038|M.thermoresistible__bu VTDLAAAVCGGIGLAASGNIDATGTNPSMFEPVHGSAPDIAGTGAADPTA
Mflv_4233|M.gilvum_PYR-GCK ITDLAAAVCGGIGLAASGNIDATLTNPSMFEPVHGSAPDIAGQGIADPTA
Mvan_2129|M.vanbaalenii_PYR-1 ITDLAAAVCGGIGLAASGNIDATLTHPSMFEPVHGSAPDIAGQGIADPTA
MSMEG_1986|M.smegmatis_MC2_155 LSDLGPACTGTIGVAPSANINPERTFPSLFEPVHGSAPDIAGQGVANPVG
MAB_4419|M.abscessus_ATCC_1997 LSDLAAAITGGLGLAASGNINPERTAISMFESVHGSAPDIAGQGIANPVA
::**..* * :.:*.*.**: : *:**.********** * *:*..
MLBr_01691|M.leprae_Br4923 AIMSLALLLAHLGEDEPAARLDQAVASYLATR------GNGRFSTGEVGE
MAV_3845|M.avium_104 AIMSVALLLAHLGEDAAATRVDRAVERYLATR------GNERPATTEVGE
Mb3019c|M.bovis_AF2122/97 AIMSVALLLSHLGEHDAAARVDRAVEAHLATR------GSERLATSDVGE
Rv2995c|M.tuberculosis_H37Rv AIMSVALLLSHLGEHDAAARVDRAVEAHLATR------GSERLATSDVGE
MMAR_1716|M.marinum_M AIMSVSLLLAHLGLDDAASRVDRAVEGYLATR------GNERLATAAVGE
MUL_1953|M.ulcerans_Agy99 AIMSVSLLLAHLGLDDTASRVDRAVEGYLATR------GNERLATAAVGE
TH_2038|M.thermoresistible__bu AIMSVALLLAHVGESDAAARVDRAVAEHLATR------GDARMSTTEIGD
Mflv_4233|M.gilvum_PYR-GCK AIMSVSLLLAHMAEIDAAARVDKAVAEHLATR------GDEKLSTTQVGD
Mvan_2129|M.vanbaalenii_PYR-1 AVMSVSLLLAHMAEVDAAARVDKAVAEHLATR------GDEKLSTAAVGE
MSMEG_1986|M.smegmatis_MC2_155 QIWCASMMLEHLGERTAAKAVLDAIEAVLADGAVLTPDLGGTATTTALGT
MAB_4419|M.abscessus_ATCC_1997 QILAGAMLLDHLNEPATANKIRLAVDQVLSDGRVRTPDLGGTATTEQLGT
: . :::* *: .* : *: *: . :* :*
MLBr_01691|M.leprae_Br4923 RIAAAL-----------
MAV_3845|M.avium_104 RIAAAL-----------
Mb3019c|M.bovis_AF2122/97 RIAAAL-----------
Rv2995c|M.tuberculosis_H37Rv RIAAAL-----------
MMAR_1716|M.marinum_M RIAAAL-----------
MUL_1953|M.ulcerans_Agy99 RIAAAL-----------
TH_2038|M.thermoresistible__bu RIAAKL-----------
Mflv_4233|M.gilvum_PYR-GCK RILGKL-----------
Mvan_2129|M.vanbaalenii_PYR-1 RILGKL-----------
MSMEG_1986|M.smegmatis_MC2_155 AIADSVGASASASAGTR
MAB_4419|M.abscessus_ATCC_1997 AIAEAAR----------
*