For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KLAVIAGDGIGPEVIGEALRVLDAVVPGVEKTEYDLGARLYHRTGEVLPDSVLDELKGHDAILLGAIGDP SMPSGVLERGLLLRIRFELDHHINLRPGRLYPGVQSPLAGNPEIDFVVVREGTEGPYTGNGGAIRVGTPH EIATEVSVNTAYGVRRVVQDAFKRAQQRRKHLTLVHKNNVLTNAGSLWWRTVQAVAAEYPEVEVAYQHVD AATIHMVTDPGRFDVIVTDNLFGDIITDLAAAVCGGIGLAASGNIDATLTNPSMFEPVHGSAPDIAGQGI ADPTAAIMSVSLLLAHMAEIDAAARVDKAVAEHLATRGDEKLSTTQVGDRILGKL*
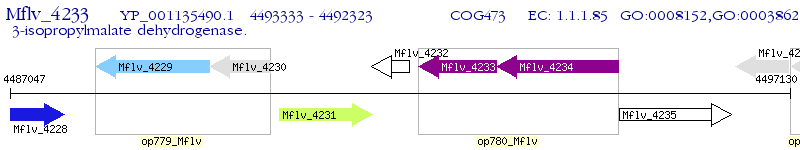
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4233 | - | - | 100% (336) | 3-isopropylmalate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3019c | leuB | 1e-161 | 83.04% (336) | 3-isopropylmalate dehydrogenase |
| M. tuberculosis H37Rv | Rv2995c | leuB | 1e-161 | 83.04% (336) | 3-isopropylmalate dehydrogenase |
| M. leprae Br4923 | MLBr_01691 | leuB | 1e-155 | 78.87% (336) | 3-isopropylmalate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3303c | - | 1e-154 | 80.36% (331) | 3-isopropylmalate dehydrogenase |
| M. marinum M | MMAR_1716 | leuB | 1e-162 | 82.89% (339) | 3-isopropylmalate dehydrogenase LeuB |
| M. avium 104 | MAV_3845 | leuB | 1e-159 | 81.85% (336) | 3-isopropylmalate dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2379 | leuB | 1e-162 | 83.73% (332) | 3-isopropylmalate dehydrogenase |
| M. thermoresistible (build 8) | TH_2038 | leuB | 1e-164 | 83.63% (336) | PROBABLE 3-ISOPROPYLMALATE DEHYDROGENASE LEUB (BETA-IPM |
| M. ulcerans Agy99 | MUL_1953 | leuB | 1e-161 | 82.01% (339) | 3-isopropylmalate dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_2129 | - | 0.0 | 93.75% (336) | 3-isopropylmalate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4233|M.gilvum_PYR-GCK ----MKLAVIAGDGIGPEVIGEALRVLDAVVPGVEKTEYDLGARLYHRTG
Mvan_2129|M.vanbaalenii_PYR-1 ----MRLAVIAGDGIGPEVITEALKVLDAVLPGVEKTEYDLGARQYHKTG
MLBr_01691|M.leprae_Br4923 ----MKLAIIGGDGIGPEVVAQAVKILDVVLPGVQKTTYDLGARRYHTTG
MAV_3845|M.avium_104 ----MKLAVIGGDGIGPEVTAEALKVLDAVLPGVDKTEYDLGARRYHATG
Mb3019c|M.bovis_AF2122/97 ----MKLAIIAGDGIGPEVTAEAVKVLDAVVPGVQKTSYDLGARRFHATG
Rv2995c|M.tuberculosis_H37Rv ----MKLAIIAGDGIGPEVTAEAVKVLDAVVPGVQKTSYDLGARRFHATG
MMAR_1716|M.marinum_M ----MKLAIVAGDGIGPEVVAQAVKVLDVVQPGVEKTNYDLGARRFHATG
MUL_1953|M.ulcerans_Agy99 ----MKLAIVAGDGIGPEVVAQAVKILDVVQPGVEKTNYDLGARRFHATG
TH_2038|M.thermoresistible__bu --VTMKLAIIPGDGIGPEVIGEAVKVLDAVLPDVDKTHFDLGARRFHATG
MSMEG_2379|M.smegmatis_MC2_155 MSSNLKLAVIAGDGIGPEVIAEAVTVLDAVLPGVEKTEYDLGARRYHATG
MAB_3303c|M.abscessus_ATCC_199 ---MTKLAIIAGDGIGPEVIGEAVKVLDAVLPGVDKTEYDLGARRYLATG
:**:: ******** :*: :**.* *.*:** :***** : **
Mflv_4233|M.gilvum_PYR-GCK EVLPDSVLDELKGHDAILLGAIGDPSMPSGVLERGLLLRIRFELDHHINL
Mvan_2129|M.vanbaalenii_PYR-1 EVLPDSVLEELKGHDAILLGAIGDPSVPSGLLERGLLLRIRFELDHHINL
MLBr_01691|M.leprae_Br4923 ELLPESVLAELREHDAILLGAVGDPSVPSGVLERGLLLRLRFELDHHINL
MAV_3845|M.avium_104 ELLPDSVIDELRAHDAILLGAIGDPSVPSGVLERGLLLRLRFELDHHINL
Mb3019c|M.bovis_AF2122/97 EVLPDSVVAELRNHDAILLGAIGDPSVPSGVLERGLLLRLRFELDHHINL
Rv2995c|M.tuberculosis_H37Rv EVLPDSVVAELRNHDAILLGAIGDPSVPSGVLERGLLLRLRFELDHHINL
MMAR_1716|M.marinum_M EILPDSVIAELREHDAILLGAIGDPSVPSGVLERGLLLRLRFELDHHINL
MUL_1953|M.ulcerans_Agy99 EILPDSVIAELREHDAILLGAIGDPSVPSGVLERGLLLRLRFELDYHINL
TH_2038|M.thermoresistible__bu EVLPDSVIDELRAHDAILLGAIGDPSVPSGVLERGLLLRIRFELDHHINL
MSMEG_2379|M.smegmatis_MC2_155 ELLSDETIEELRGYDAILLGAIGDPSVPSGVLERGMLLKLRFALDHHVNL
MAB_3303c|M.abscessus_ATCC_199 ELMPDSVLDELRGHDAILLGAIGDPAVPSGVLERGLLLNTRFALDHHINL
*::.:..: **: :*******:***::***:****:**. ** **:*:**
Mflv_4233|M.gilvum_PYR-GCK RPGRLYPGVQSPLA---GNPEIDFVVVREGTEGPYTGNGGAIRVGTPHEI
Mvan_2129|M.vanbaalenii_PYR-1 RPARLYPGVQSPLA---GNPEIDFVVVREGTEGPYTGNGGAIRVGTPHEI
MLBr_01691|M.leprae_Br4923 RPARLYPGVNSLLA---GKPDIDFVVVREGTEGPYTGTGGAIRVGTPNEV
MAV_3845|M.avium_104 RPGRLYPGVSSPLA---GNPDIDFVVVREGTEGPYTGTGGAIRVGTPNEV
Mb3019c|M.bovis_AF2122/97 RPARLYPGVASPLS---GNPGIDFVVVREGTEGPYTGNGGAIRVGTPNEV
Rv2995c|M.tuberculosis_H37Rv RPARLYPGVASPLS---GNPGIDFVVVREGTEGPYTGNGGAIRVGTPNEV
MMAR_1716|M.marinum_M RPGRLYPGVKSPLALEPGNPEIDFVVVREGTEGPYTGNGGAIRVGTANEV
MUL_1953|M.ulcerans_Agy99 RPGRLYPGVKSPLALEPGNPEIDFVVVREGTEGPYTGNGGAIRVGTANEV
TH_2038|M.thermoresistible__bu RPARLYEGVSSPLA---GNPDIDFVVVREGTEGPYTGTGGAIRVGTPNEV
MSMEG_2379|M.smegmatis_MC2_155 RPGRLYPGVTGPLV---GNPPIDFVVVREGTEGPYTGNGGALRVGTPHEV
MAB_3303c|M.abscessus_ATCC_199 RPAKLYPGVTGPLA---GNPAIDLVVVREGTEGPYTGNGGAIRVDTPHEV
**.:** ** . * *:* **:*************.***:**.*.:*:
Mflv_4233|M.gilvum_PYR-GCK ATEVSVNTAYGVRRVVQDAFKRAQQRRKHLTLVHKNNVLTNAGSLWWRTV
Mvan_2129|M.vanbaalenii_PYR-1 ATEVSVNTAFGVRRAVQDAFGRAQQRRKHLTLVHKNNVLTNAGSLWWRTV
MLBr_01691|M.leprae_Br4923 ATEVSVNTAFGVRRVVQDAFERARQRRKHLTLVHKNNVLTYAGTLWCRIV
MAV_3845|M.avium_104 ATEVSQNTAFGVRRVVVDAFERARRRRKHLTLVHKTNVLTFAGKLWSRIV
Mb3019c|M.bovis_AF2122/97 ATEVSVNTAFGVRRVVADAFERARRRRKHLTLVHKTNVLTLAGGLWLRTV
Rv2995c|M.tuberculosis_H37Rv ATEVSVNTAFGVRRVVADAFERARRRRKHLTLVHKTNVLTFAGGLWLRTV
MMAR_1716|M.marinum_M ATEVSVNTAFGVRRVVRDAFERAMRRRKHLTLVHKNNVLTFAGSLWWRTV
MUL_1953|M.ulcerans_Agy99 ATEVSVNTAFGVRRVVRDAFERAMRRRKHLTLVHKNNVLTFAGSLWWRTV
TH_2038|M.thermoresistible__bu ATEVSLNTAFGVRRVVENAFQRAAQRRKHLTLVHKTNVLTYAGALWSRTV
MSMEG_2379|M.smegmatis_MC2_155 ATEVSVNTAFGVERVVRDAFARAQDRRKHLTLVHKTNVLAFAGSLWSRTV
MAB_3303c|M.abscessus_ATCC_199 ATEVSLNTRFGVERVVRNAFERAEARSKNLTLVHKNNVLKYAGTLWTRTV
***** ** :**.*.* :** ** * *:******.*** ** ** * *
Mflv_4233|M.gilvum_PYR-GCK QAVAAEYPEVEVAYQHVDAATIHMVTDPGRFDVIVTDNLFGDIITDLAAA
Mvan_2129|M.vanbaalenii_PYR-1 QTVAAEYPDVEVAYQHVDAATIHMVTDPGRFDVIVTDNLFGDIITDLAAA
MLBr_01691|M.leprae_Br4923 QEVGEKYPDVEVVYQHIDAATIYLVTEPSRFDVIVTDNLFGDIITDLAAA
MAV_3845|M.avium_104 AEVGRDYPDVEVAYQHIDAATIFMVTDPGRFDVIVTDNLFGDIITDLSAA
Mb3019c|M.bovis_AF2122/97 DEVGECYPDVEVAYQHVDAATIHMITDPGRFDVIVTDNLFGDIITDLAAA
Rv2995c|M.tuberculosis_H37Rv DEVGECYPDVEVAYQHVDAATIHMITDPGRFDVIVTDNLFGDIITDLAAA
MMAR_1716|M.marinum_M QEIGEEYPDVELAYQHVDAATIHMVTDPGRFDVIVTDNLFGDIITDLAAA
MUL_1953|M.ulcerans_Agy99 QEIGEEYPDVELAYQHVDAATIHMVTDPGRFDVIVTDNLFGDIITDLAAA
TH_2038|M.thermoresistible__bu QEVGEQYPDVEVAYQHVDAATIHLVTDPGRFDVIVTDNLFGDIVTDLAAA
MSMEG_2379|M.smegmatis_MC2_155 DRVAQEYPDVEVAYQHIDAATIHMVTDPGRFDVIVTDNLFGDIITDLAAA
MAB_3303c|M.abscessus_ATCC_199 NEVAAEYPDVQVSYQHIDAATIHMVTDPGRFDVIVTDNLFGDIITDLTAA
:. **:*:: ***:*****.::*:*.**************:***:**
Mflv_4233|M.gilvum_PYR-GCK VCGGIGLAASGNIDATLTNPSMFEPVHGSAPDIAGQGIADPTAAIMSVSL
Mvan_2129|M.vanbaalenii_PYR-1 VCGGIGLAASGNIDATLTHPSMFEPVHGSAPDIAGQGIADPTAAVMSVSL
MLBr_01691|M.leprae_Br4923 VCGGIALAASGNIDATRTNPSMFEPVHGSAPDIAGQGIADPTAAIMSLAL
MAV_3845|M.avium_104 VCGGIGLAASGNIDGTRTNPSMFEPVHGSAPDIAGQGVADPTAAIMSVAL
Mb3019c|M.bovis_AF2122/97 VCGGIGLAASGNIDATRANPSMFEPVHGSAPDIAGQGIADPTAAIMSVAL
Rv2995c|M.tuberculosis_H37Rv VCGGIGLAASGNIDATRANPSMFEPVHGSAPDIAGQGIADPTAAIMSVAL
MMAR_1716|M.marinum_M VCGGIGLAASGNIDATRTNPSMFEPVHGSAPDIAGQGIADPTAAIMSVSL
MUL_1953|M.ulcerans_Agy99 VCGGIGLAASGNIDATRTNPSMFEPVHGSAPDIAGQGIADPTAAIMSVSL
TH_2038|M.thermoresistible__bu VCGGIGLAASGNIDATGTNPSMFEPVHGSAPDIAGTGAADPTAAIMSVAL
MSMEG_2379|M.smegmatis_MC2_155 VCGGIGLAASGNIDATRTNPSMFEPVHGSAPDIAGQGIADPTAAVMSVGL
MAB_3303c|M.abscessus_ATCC_199 VTGGIGLAASGNIDGSGKNPSMFEPVHGSAPDIAGQGIADPTAAIVSVSL
* ***.********.: :**************** * ******::*:.*
Mflv_4233|M.gilvum_PYR-GCK LLAHMAEIDAAARVDKAVAEHLATRGDEKLSTTQVGDRILGKL
Mvan_2129|M.vanbaalenii_PYR-1 LLAHMAEVDAAARVDKAVAEHLATRGDEKLSTAAVGERILGKL
MLBr_01691|M.leprae_Br4923 LLAHLGEDEPAARLDQAVASYLATRGNGRFSTGEVGERIAAAL
MAV_3845|M.avium_104 LLAHLGEDAAATRVDRAVERYLATRGNERPATTEVGERIAAAL
Mb3019c|M.bovis_AF2122/97 LLSHLGEHDAAARVDRAVEAHLATRGSERLATSDVGERIAAAL
Rv2995c|M.tuberculosis_H37Rv LLSHLGEHDAAARVDRAVEAHLATRGSERLATSDVGERIAAAL
MMAR_1716|M.marinum_M LLAHLGLDDAASRVDRAVEGYLATRGNERLATAAVGERIAAAL
MUL_1953|M.ulcerans_Agy99 LLAHLGLDDTASRVDRAVEGYLATRGNERLATAAVGERIAAAL
TH_2038|M.thermoresistible__bu LLAHVGESDAAARVDRAVAEHLATRGDARMSTTEIGDRIAAKL
MSMEG_2379|M.smegmatis_MC2_155 LLSHVGQTEAAARVDKAVSEHLATRGDEKLSTSAVGERIRSLL
MAB_3303c|M.abscessus_ATCC_199 LLDHLGETEAAARVERAVEQHLATRGEEKLSTSATGDRIASFL
** *:. .*:*:::** :*****. : :* *:** . *