For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SLAQPLDVTRPRPDIAVLTLNRPDKLNALSYELVEALHAALDDIAADNDCRVVVLTGAGRGFCSGLDLSA PNPPEAAGGTEFPRSGMRWQERIANLTAKLHRLRQPVIAAVNGPAYGGGFALAAASDIRLAAESARFCTQ FIKLGIGGCDIGVSYTLPRIIGAGPAFDLILTARAVDATEALRLGLVSRVCADVTAEAMDIAETLCGYGK FGVESTKQVLWANLDAGCLEAALHVENRSQILASTEGGVRAFSEEFRRRPR*
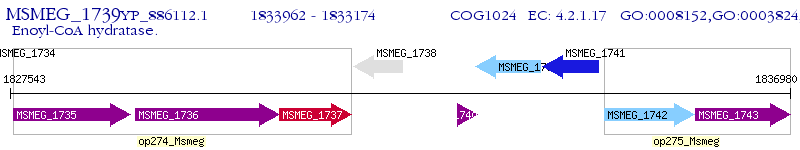
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1739 | - | - | 100% (262) | enoyl-CoA hydratase/isomerase family protein |
| M. smegmatis MC2 155 | MSMEG_3139 | - | 2e-49 | 41.90% (253) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_2229 | - | 2e-27 | 37.50% (200) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1507 | echA12 | 4e-50 | 43.14% (255) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_4817 | - | 1e-110 | 77.13% (258) | enoyl-CoA hydratase/isomerase |
| M. tuberculosis H37Rv | Rv1472 | echA12 | 2e-49 | 42.75% (255) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_01241 | - | 4e-47 | 41.20% (233) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_2737c | - | 5e-53 | 44.49% (236) | enoyl-CoA hydratase |
| M. marinum M | MMAR_2278 | echA12 | 3e-50 | 42.91% (254) | enoyl-CoA hydratase, EchA12 |
| M. avium 104 | MAV_3942 | - | 8e-51 | 41.98% (262) | enoyl-CoA hydratase/isomerase |
| M. thermoresistible (build 8) | TH_2798 | - | 4e-54 | 46.19% (236) | PUTATIVE Enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_1480 | echA12 | 6e-50 | 42.52% (254) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_1625 | - | 1e-114 | 77.99% (259) | enoyl-CoA hydratase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1507|M.bovis_AF2122/97 --MPHRCAAQVVAGYRSTVSLVLVEHPRPEIAQITLNRPERMNSMAFDVM
Rv1472|M.tuberculosis_H37Rv --MPHRCAAQVVAGYRSTVSLVLVEHPRPEIAQITLNRPERMNSMAFDVM
MMAR_2278|M.marinum_M ------------------MALVLLEHPRPDIALVTLNRPERMNSMAFDVM
MUL_1480|M.ulcerans_Agy99 ------------------MSLVLLEHPRPDIALVTLNRPERMNSIAFDVM
TH_2798|M.thermoresistible__bu ---------------------VLVDHPRPHIALVTLNRPERMNSMAFDVM
MAB_2737c|M.abscessus_ATCC_199 ------------------MSFVLVDRPRPEIALVTLNRPERMNAMAFDVM
MLBr_01241|M.leprae_Br4923 MSQTDASCTIAELPYRSVTDLVVLDFPRPEVALITLNRPGRMNSMALDLM
Mflv_4817|M.gilvum_PYR-GCK -----------------MSSVLEVARPTPEIAVLTLNRPDKLNALSYELV
Mvan_1625|M.vanbaalenii_PYR-1 -----------------MSSVVEVTRPLPEITVVTLNRPDKLNALSYELV
MSMEG_1739|M.smegmatis_MC2_155 ---------------MSLAQPLDVTRPRPDIAVLTLNRPDKLNALSYELV
MAV_3942|M.avium_104 ---------------MTMPSTLELAAPGDGVRVLRLNRPHRLNAIDEAMQ
: : * : : **** ::*:: :
Mb1507|M.bovis_AF2122/97 VPLKEALAQVSYDNSVRVVVLTGAGRGFSSGADHKSAGVVPHVENLTRPT
Rv1472|M.tuberculosis_H37Rv VPLKEALAQVSYDNSVRVVVLTGAGRGFSPGADHKSAGVVPHVENLTRPT
MMAR_2278|M.marinum_M VPLKEVLEEVSYDNSVRVVVLTGAGRGFSSGADHKSAGAVPNVGGLTRPT
MUL_1480|M.ulcerans_Agy99 VPLKEVLEEVSYDNSVRVVVLTGAGRGFSSGADHKSAGAVPNVGGLTRPT
TH_2798|M.thermoresistible__bu VPLKAALDEITYDNDVRVVVLTGAGRGFSSGADHKSAGTVPHVDGLTRPS
MAB_2737c|M.abscessus_ATCC_199 LPFKQMLVDISHDNDVRAVVITGAGKGFCSGADQKSAGPIPHIGGLTQPT
MLBr_01241|M.leprae_Br4923 KSLKQVLKRITYDHSVRVVVLTGAGRGFCSGADQKFTAPVPQVEGLTQPV
Mflv_4817|M.gilvum_PYR-GCK EALHAELDALARDNACRAVVLTGAGRGFCSGLDLSAPNPEETGGGTEFPR
Mvan_1625|M.vanbaalenii_PYR-1 ETLHDALDAVAADNTCRVVVLTGAGRGFCSGLDLSAPNPEETGGGTEFPR
MSMEG_1739|M.smegmatis_MC2_155 EALHAALDDIAADNDCRVVVLTGAGRGFCSGLDLSAPNPPEAAGGTEFPR
MAV_3942|M.avium_104 TELRLVLDDLAADHAVRAVVLTGAGRGFCAGIDMRDFGPSMVGADAPALE
:: * :: *: *.**:****:**..* * .
Mb1507|M.bovis_AF2122/97 YALRSMELLDDVILMLRRLHQPVIAAVNGPAIGGGLCLALAADIRVASSS
Rv1472|M.tuberculosis_H37Rv YALRSMELLDDVILMLRRLHQPVIAAVNGPAIGGGLCLALAADIRVASSS
MMAR_2278|M.marinum_M YALRSMELLDEVILMLRRLHQPVIAAVNGPAIGGGLCLALAADVRVAAAS
MUL_1480|M.ulcerans_Agy99 YALRSMELLDEVILMLRRLHQPVIAAVNGPAIGGGLCLALAADVRVAAAS
TH_2798|M.thermoresistible__bu YGLRSMEVLDDVILTLRKLHQPVIAAVNGAAIGGGLCLALAADIRIASTE
MAB_2737c|M.abscessus_ATCC_199 IALRSMELLDEVILTLRRMHQPVIAAINGAAIGGGLCLALACDVRVASQD
MLBr_01241|M.leprae_Br4923 RALRAMELLEEVILALRRLHQPVIAAINGPAIGGGLCLALAADVRVASTR
Mflv_4817|M.gilvum_PYR-GCK SGMRWQERIADLTAKIHRLRQPVIAAVQGVAYGGGLGIAAACDIRVAAPT
Mvan_1625|M.vanbaalenii_PYR-1 SGMRWQERIADLTAKLHRLRQPAIAAVNGVAYGGGLGIAAACDIRVASPS
MSMEG_1739|M.smegmatis_MC2_155 SGMRWQERIANLTAKLHRLRQPVIAAVNGPAYGGGFALAAASDIRLAAES
MAV_3942|M.avium_104 R-MRFQERMAALAEAVWALPQPVIAAVNGPCVGAGLALCLAADIRICSAA
:* * : : : : **.***::* . *.*: :. *.*:*:.:
Mb1507|M.bovis_AF2122/97 AYFRAAGINNGLTASELGLSYLLPRAIGSSRAFEIMLTGRDVSAEEAERI
Rv1472|M.tuberculosis_H37Rv AYFRAAGINNGLTASELGLSYLLPRAIGSSRAFEIMLTGRDVSAEEAERI
MMAR_2278|M.marinum_M AYFRAAGINNGLTASELGLSYLLPRAIGSSRAFEIMLTGRDVTAEEAERI
MUL_1480|M.ulcerans_Agy99 AYFRAAGINNGLTASELGLSYLLPRAIGSSRAFEIMLTGRDVTAEEAERI
TH_2798|M.thermoresistible__bu AYFRAAGINNGLTASELGLSYLLPRAIGSSRAFEIMLTGRDVDAEEAERI
MAB_2737c|M.abscessus_ATCC_199 AYFRAAGINNGLTASELGLSYLLPRAIGTSRASDIMLTGRDVDADEAERI
MLBr_01241|M.leprae_Br4923 AYFRAAGINNGLSASELGLSYLLPRAVGSSRAFEIMLSGRDVGAEEAEQI
Mflv_4817|M.gilvum_PYR-GCK ARFCTQFIKLGLGGCDIGVSYTLPRIVGAGVAFDMILTARTVAADEALRL
Mvan_1625|M.vanbaalenii_PYR-1 ARFCTQFIKLGLGGCDIGVSYTLPRILGAGPAFDMILTARAVDADEALRL
MSMEG_1739|M.smegmatis_MC2_155 ARFCTQFIKLGIGGCDIGVSYTLPRIIGAGPAFDLILTARAVDATEALRL
MAV_3942|M.avium_104 ATFANAAILLGLSGAEMGMSYHLPRIVGTGVAADWMLTGRTVSAAEADRR
* * * *: ..::*:** *** :*:. * : :*:.* * * ** :
Mb1507|M.bovis_AF2122/97 GLVSRQVPDEQLLDACYAIAARMAGFSRPGIELTKRTLWSGLDAASLEAH
Rv1472|M.tuberculosis_H37Rv GLVSRQVPDEQLLDACYAIAARMAGFSRPGIELTKRTLWSGLDAASLEAH
MMAR_2278|M.marinum_M GLVSCRVPDGQLLDTCYAIAARMAAFSRPGIELTKRTLWSGLDAASLEGH
MUL_1480|M.ulcerans_Agy99 GLVSCRVPDGQLLDTCYAITARMAAFSRPGIELTKRTLWSGLDAASLEGH
TH_2798|M.thermoresistible__bu GLVSRRVPPAELLDTCYELADRIAGFSRPGTELTKRTLWSGLDAGSLESH
MAB_2737c|M.abscessus_ATCC_199 GLVSRKVASESLLEECYAIGERIAGFSRPGIELTKRTIWSGLDAASLESH
MLBr_01241|M.leprae_Br4923 GLVSYRVSDDRLLDTCYSIAARMATFSRSGTELTKRALWGGLDAASLDKH
Mflv_4817|M.gilvum_PYR-GCK GLVSRVS--DTPVDEALAIAETLCNYGKFGVESTKQVLWANLVASSLEAA
Mvan_1625|M.vanbaalenii_PYR-1 GLVSRVA--DSALDEALAVAETLCGFGKFGVESTKQVLWANLEASSLEAA
MSMEG_1739|M.smegmatis_MC2_155 GLVSRVC--ADVTAEAMDIAETLCGYGKFGVESTKQVLWANLDAGCLEAA
MAV_3942|M.avium_104 GLVSEVVEPDRLVERAVELARLIAGHAPLGAQLTKRALQVNTDAASLASA
**** . : :. .. * : **:.: . *..*
Mb1507|M.bovis_AF2122/97 MQAEGLGQLFVRLLTANFEEAVAARAEQRAPVFTDDT-------
Rv1472|M.tuberculosis_H37Rv MQAEGLGQLFVRLLTANFEEAVAARAEQRAPVFTDDT-------
MMAR_2278|M.marinum_M MQAEGLGQLFVRLLTANFEEAVAARAEKRPAVFTDDK-------
MUL_1480|M.ulcerans_Agy99 MQAEGLGQLFVRLLTANFEEAVAARAEKRPAVFTDDK-------
TH_2798|M.thermoresistible__bu MHAEGLGQLYIRLLTHNFEEAVAARKEKRAPRFPDDKI------
MAB_2737c|M.abscessus_ATCC_199 MHQEGLGQLYVRLLTDNFEEATAARKEKRPAEFRDKR-------
MLBr_01241|M.leprae_Br4923 MQSESLAQLFIALHTSNFEEAAAPCTEKRPTVLVDARGCATSPG
Mflv_4817|M.gilvum_PYR-GCK LHLENRSQILA---STSGEMRELAAKILRKG-------------
Mvan_1625|M.vanbaalenii_PYR-1 LHLENRSQILA---STSGEMREAAAKILRKP-------------
MSMEG_1739|M.smegmatis_MC2_155 LHVENRSQILA---STEGGVRAFSEEFRRRPR------------
MAV_3942|M.avium_104 MELENRNQVIA---HATDEAAARRQKWSR---------------
:. *. *: *