For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STPHHGRHELGQNFLSDRRVIADIVEIVSRTNGPIIEIGAGDGALTIPLQRLARPLTAVEVDARRARRLA QRTARSAPGPASRPTEVVAADFLRYPLPRSPHVVVGNLPFHLTTAILRRLLHGPGWTTAVLLMQWEVARR RAAVGGATMMTAQWWPWFEFGLARKVSAASFTPRPAVDAGLLTITRRSRPLVDVADRARYQALVHRVFTG RGHGMAQIRQRLPTPVPRTWLRANGIAPNSLPRQLSAAQWAALFEQTRLTGAQRVDRPRDVQHGRAHRRR GGEVDRPATHHKQTGPVVGQRQPQRGRDADADPDDQRTAPPVTRHHQGERRDEDQADHQDRPLTGEHLAG EFLWRHASFDSSASTTLVSRKARVNGPTPPGLGDT*
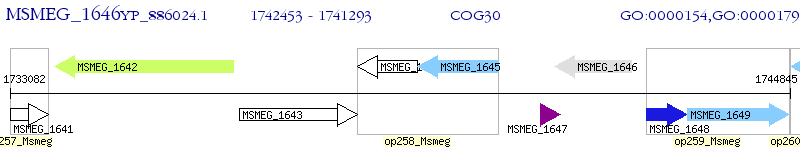
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1646 | - | - | 100% (386) | ribosomal RNA adenine dimethylase family protein |
| M. smegmatis MC2 155 | MSMEG_5438 | ksgA | 4e-12 | 31.91% (188) | dimethyladenosine transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1037 | ksgA | 1e-15 | 35.19% (216) | dimethyladenosine transferase |
| M. gilvum PYR-GCK | Mflv_1933 | ksgA | 1e-12 | 31.91% (188) | dimethyladenosine transferase |
| M. tuberculosis H37Rv | Rv1010 | ksgA | 1e-15 | 35.19% (216) | dimethyladenosine transferase |
| M. leprae Br4923 | MLBr_00241 | ksgA | 7e-15 | 32.09% (215) | dimethyladenosine transferase |
| M. abscessus ATCC 19977 | MAB_1131 | ksgA | 2e-12 | 33.16% (187) | dimethyladenosine transferase |
| M. marinum M | MMAR_4478 | ksgA | 6e-11 | 33.96% (159) | dimethyladenosine transferase KsgA |
| M. avium 104 | MAV_1148 | ksgA | 2e-14 | 34.92% (189) | dimethyladenosine transferase |
| M. thermoresistible (build 8) | TH_0980 | ksgA | 2e-10 | 31.72% (186) | PROBABLE DIMETHYLADENOSINE TRANSFERASE KSGA |
| M. ulcerans Agy99 | MUL_4650 | ksgA | 4e-11 | 33.96% (159) | dimethyladenosine transferase |
| M. vanbaalenii PYR-1 | Mvan_4800 | ksgA | 8e-13 | 31.91% (188) | dimethyladenosine transferase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4478|M.marinum_M --------------------------------------------------
MUL_4650|M.ulcerans_Agy99 --------------------------------------------------
Mb1037|M.bovis_AF2122/97 ------MCCTSGCALTIRLLGRTEIRRLAKELDFRPRKSLGQNFVHDANT
Rv1010|M.tuberculosis_H37Rv ------MCCTSGCALTIRLLGRTEIRRLAKELDFRPRKSLGQNFVHDANT
MLBr_00241|M.leprae_Br4923 ------MQRKSGCTLTIRLLERTEIRWLVKELECRPRKSLGQNFVHDANT
MAV_1148|M.avium_104 ---MASVQRKSWCALTIRLLGRTEIRRLAKELEFRPRKSLGQNFVHDANT
Mflv_1933|M.gilvum_PYR-GCK --------------MTIRLLGRTEIRHLAKSIDFRPRKSFGQNFVHDANT
Mvan_4800|M.vanbaalenii_PYR-1 --------------MTIRLLGRTEIRHLAKSIDFRPRKSFGQNFVHDANT
TH_0980|M.thermoresistible__bu --------------------------------------------------
MAB_1131|M.abscessus_ATCC_1997 MGRLAGMQRESWCPLTIRLLGRTEIRQLAKDLDFRPTKTLGQNFVHDANT
MSMEG_1646|M.smegmatis_MC2_155 -----------------------------MSTPHHGRHELGQNFLSDRRV
MMAR_4478|M.marinum_M MRKVVTVSGITRSDQVLEVGPGLGSLTLALLERGAAVTAVEIDPVLAARL
MUL_4650|M.ulcerans_Agy99 MRKVVAVSGTTRSDQVLEVGPGLGSLTLALLERGAAVTAVEIDPVLAARL
Mb1037|M.bovis_AF2122/97 VRRVVAASGVSRSDLVLEVGPGLGSLTLALLDRGATVTAVEIDPLLASRL
Rv1010|M.tuberculosis_H37Rv VRRVVAASGVSRSDLVLEVGPGLGSLTLALLDRGATVTAVEIDPLLASRL
MLBr_00241|M.leprae_Br4923 VRRVVSTSRVNRSDFVLEVGPGFGSLTLALLDCGAAVSAIEIDPVLAGRL
MAV_1148|M.avium_104 VRRVVSTSGVSRSDHVLEVGPGLGSLTLALLDRGAHVTAVEIDPVLAERL
Mflv_1933|M.gilvum_PYR-GCK VRRIVSASSVNRSDHVLEVGPGLGSLTLALLDRGARVTAVEIDPVLATQL
Mvan_4800|M.vanbaalenii_PYR-1 VRRIVSASSINRSDHVLEVGPGLGSLTLALLDRGAKVTAVEIDPVLANQL
TH_0980|M.thermoresistible__bu VRRIVSASGINKQDHVLEVGPGLGSLTLGLLDRGAHVTAVEIDSVLARQL
MAB_1131|M.abscessus_ATCC_1997 IRRIVSASGVGKGDHVIEVGPGLGSLTLGLLDKGAAVTAVEVDPVLARRL
MSMEG_1646|M.smegmatis_MC2_155 IADIVEIVSRTNG-PIIEIGAGDGALTIPLQRLARPLTAVEVDARRARRL
: :* . ::*:*.* *:**: * . ::*:*:*. * :*
MMAR_4478|M.marinum_M PQTVAEHSDSEIGR-LTVVHHDILTLRRQDVADQPTAVVANLPYNVAVPA
MUL_4650|M.ulcerans_Agy99 PQTVAEHSDSEIGR-LTVVHHDILTLRRQDVADQPTAVVANLPYNVAVPA
Mb1037|M.bovis_AF2122/97 QQTVAEHSHSEVHR-LTVVNRDVLALRREDLAAAPTAVVANLPYNVAVPA
Rv1010|M.tuberculosis_H37Rv QQTVAEHSHSEVHR-LTVVNRDVLALRREDLAAAPTAVVANLPYNVAVPA
MLBr_00241|M.leprae_Br4923 PQTVAEHSNNEIHR-LTVCNRDVLSFRRGDLATEPTALVANLPYNVAVPA
MAV_1148|M.avium_104 PHTVAEHSHSEIQR-LTVLNRDVLTLRRDELAEPPTAVVANLPYNVAVPA
Mflv_1933|M.gilvum_PYR-GCK PTTIAAHSHSEVNR-LTVLNRDILTFKQSDMTEMPTALVANLPYNVAVPA
Mvan_4800|M.vanbaalenii_PYR-1 PTTIAAHSHSEVNR-LTVLNRDILTFKQSDMTEMPTALVANLPYNVAVPA
TH_0980|M.thermoresistible__bu PKTIADHSHSEINR-LIVLNRDILDLYPQDLETQPTALVANLPYNVAVPA
MAB_1131|M.abscessus_ATCC_1997 PTTIKEHSHSEFNR-FSVIHQDVLNLTPADIQGEPTAMVANLPYNVAVPA
MSMEG_1646|M.smegmatis_MC2_155 AQRTARSAPGPASRPTEVVAADFLRYP---LPRSPHVVVGNLPFHLTT-A
: . * * *.* : * .:*.***::::. *
MMAR_4478|M.marinum_M LLHLLAEFPSIRVVTVMVQAEVAERLAAEPGGKEYGVPSVKVGFYGRVRR
MUL_4650|M.ulcerans_Agy99 LLHLLAEFPSIRVVTVMVQAEVAERLAAEPGGKEYGVPSVKVGFYGRVRR
Mb1037|M.bovis_AF2122/97 LLHLLVEFPSIRVVTVMVQAEVAERLAAEPGSKEYGVPSVKLRFFGRVRR
Rv1010|M.tuberculosis_H37Rv LLHLLVEFPSIRVVTVMVQAEVAERLAAEPGSKEYGVPSVKLRFFGRVRR
MLBr_00241|M.leprae_Br4923 LLHLLAEFPSIRTVTVMVQAEVAERLAAEPGGKDYGVPSVKLSFFGRVRR
MAV_1148|M.avium_104 LLHLLAEFPSIRTVTVMVQAEVAERLAAEPGGKEYGVPSVKVRFFGRVRR
Mflv_1933|M.gilvum_PYR-GCK LLHLLAEFPSIRTVMVMVQAEVAERLAAEPGGKDYGVPSAKVRFFGNVRR
Mvan_4800|M.vanbaalenii_PYR-1 LLHLLAEFPSIRTVMVMVQAEVAERLAAEPGGKDYGVPSAKVRFFGNVRR
TH_0980|M.thermoresistible__bu LLHLLAEFPTIRTVMVMVQAEVAERLAAEPGSKEYGVPSAKVRFYGNVRR
MAB_1131|M.abscessus_ATCC_1997 LLRILTEFDSVKTILVMVQSEVADRLTAEPGSRIYGVPSVKVRYFGKVKR
MSMEG_1646|M.smegmatis_MC2_155 ILRRLLHGPGWTTAVLLMQWEVARRRAAVGGATMMTAQWWPWFEFGLAR-
:*: * . . :::* *** * :* *. . :* .:
MMAR_4478|M.marinum_M CGMVSPTVFWPIPRVYSGLVRIDRYE------------------------
MUL_4650|M.ulcerans_Agy99 CGMVSPTVFWPIPRVYSGLVRIDRHE------------------------
Mb1037|M.bovis_AF2122/97 CGMVSPTVFWPIPRVYSGLVRIDRYE------------------------
Rv1010|M.tuberculosis_H37Rv CGMVSPTVFWPIPRVYSGLVRIDRYE------------------------
MLBr_00241|M.leprae_Br4923 CGMVSPTVFWPIPRVYSGLVRVDRYA------------------------
MAV_1148|M.avium_104 CGMVSPTVFWPIPRVYSGLVRIDRYP------------------------
Mflv_1933|M.gilvum_PYR-GCK YGMVSPTVFWPIPRVYSGLVRIDRYE------------------------
Mvan_4800|M.vanbaalenii_PYR-1 YGMVSPTVFWPIPRVYSGLVRIDRYE------------------------
TH_0980|M.thermoresistible__bu YGMVSPTVFWPIPRVYSGLVRLDRYE------------------------
MAB_1131|M.abscessus_ATCC_1997 LGTVSPAVFWPIPRVYSGLVRIDRHA------------------------
MSMEG_1646|M.smegmatis_MC2_155 --KVSAASFTPRPAVDAGLLTITRRSRPLVDVADRARYQALVHRVFTGRG
**.: * * * * :**: : *
MMAR_4478|M.marinum_M ---------------------------TTPWPTDENFRRQVFELVDIAFA
MUL_4650|M.ulcerans_Agy99 ---------------------------TTPWPTDENFRRQVFELVDIAFA
Mb1037|M.bovis_AF2122/97 ---------------------------TSPWPTDDAFRRRVFELVDIAFA
Rv1010|M.tuberculosis_H37Rv ---------------------------TSPWPTDDAFRRRVFELVDIAFA
MLBr_00241|M.leprae_Br4923 ---------------------------TSPWPTDDAFRRQVFELVDIAFT
MAV_1148|M.avium_104 ---------------------------TSPWPTDPAFRRQVFQLVDIAFG
Mflv_1933|M.gilvum_PYR-GCK ---------------------------TSPWPTDTEFQEQVFELIDIAFA
Mvan_4800|M.vanbaalenii_PYR-1 ---------------------------TSPWPTDPEFRQQVFELIDVAFA
TH_0980|M.thermoresistible__bu ---------------------------SPKWPMDAEFRERVFELIDIAFA
MAB_1131|M.abscessus_ATCC_1997 ---------------------------DAAWPSDAGFRKELFDLIDISFA
MSMEG_1646|M.smegmatis_MC2_155 HGMAQIRQRLPTPVPRTWLRANGIAPNSLPRQLSAAQWAALFEQTRLTGA
. :*: ::
MMAR_4478|M.marinum_M QRRKTSRNAFLEWAGS--------------------GNESANRLLAASID
MUL_4650|M.ulcerans_Agy99 QRRKTSRNAFLEWAGS--------------------GNESANRLLAASID
Mb1037|M.bovis_AF2122/97 QRRKTSRNAFVQWAGS--------------------GSESANRLLAASID
Rv1010|M.tuberculosis_H37Rv QRRKTSRNAFVQWAGS--------------------GSESANRLLAASID
MLBr_00241|M.leprae_Br4923 QRRKTSRNAFVKWAGS--------------------SNESANRLLAASID
MAV_1148|M.avium_104 QRRKTCRNAFVDWAGS--------------------GNESADRLLAASID
Mflv_1933|M.gilvum_PYR-GCK QRRKTSRNAFAEWAGS--------------------GNESASRLLAASID
Mvan_4800|M.vanbaalenii_PYR-1 QRRKTSRNAFAEWAGS--------------------GNESASRLLAASID
TH_0980|M.thermoresistible__bu QRRKTSRNAFAQWAGS--------------------GNESARRLLAASID
MAB_1131|M.abscessus_ATCC_1997 QRRKTSRNAFAEWAGS--------------------GDESARRLLAASID
MSMEG_1646|M.smegmatis_MC2_155 QRVDRPRDVQHGRAHRRRGGEVDRPATHHKQTGPVVGQRQPQRGRDADAD
** . *:. * ..... * *. *
MMAR_4478|M.marinum_M PARRGETLSIEDFVR------------------------LLTRSADTSGG
MUL_4650|M.ulcerans_Agy99 PARRGETLSIEDFVR------------------------LLTRSADTSGG
Mb1037|M.bovis_AF2122/97 PARRGETLSIDDFVR------------------------LLRRSGGSDEA
Rv1010|M.tuberculosis_H37Rv PARRGETLSIDDFVR------------------------LLRRSGGSDEA
MLBr_00241|M.leprae_Br4923 PARRGETLSIDDFVR------------------------LLRRS--DGR-
MAV_1148|M.avium_104 PARRGETLSIDDFVR------------------------LLQRS--ADRG
Mflv_1933|M.gilvum_PYR-GCK PSRRGETLSINDFVR------------------------LLQRSADWHTV
Mvan_4800|M.vanbaalenii_PYR-1 PSRRGETLSINDFVR------------------------LLQRSADWHVV
TH_0980|M.thermoresistible__bu PARRGETLSIADFVR------------------------LLQRTDEVAAP
MAB_1131|M.abscessus_ATCC_1997 PARRGETLDVADFAR------------------------LLERSAIS---
MSMEG_1646|M.smegmatis_MC2_155 PDDQRTAPPVTRHHQGERRDEDQADHQDRPLTGEHLAGEFLWRHASFDSS
* : : : . : :* *
MMAR_4478|M.marinum_M PPDAGVAGDEGQVPGPVSAT----
MUL_4650|M.ulcerans_Agy99 PPDAGVAGDEGQVPGPVSAT----
Mb1037|M.bovis_AF2122/97 TSTGRDAR-APDISGHASAS----
Rv1010|M.tuberculosis_H37Rv TSTGRDAR-APDISGHASAS----
MLBr_00241|M.leprae_Br4923 --DDAAVR-SASAS----------
MAV_1148|M.avium_104 GSDREGTS-PPTAGQGAPAC----
Mflv_1933|M.gilvum_PYR-GCK PKADDAGDDADAQAKADGAQVSTL
Mvan_4800|M.vanbaalenii_PYR-1 PKPESA---ATAVATDEDAQVPTL
TH_0980|M.thermoresistible__bu AEK---------------------
MAB_1131|M.abscessus_ATCC_1997 ------------------------
MSMEG_1646|M.smegmatis_MC2_155 ASTTLVSRKARVNGPTPPGLGDT-