For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VQRKSGCTLTIRLLERTEIRWLVKELECRPRKSLGQNFVHDANTVRRVVSTSRVNRSDFVLEVGPGFGSL TLALLDCGAAVSAIEIDPVLAGRLPQTVAEHSNNEIHRLTVCNRDVLSFRRGDLATEPTALVANLPYNVA VPALLHLLAEFPSIRTVTVMVQAEVAERLAAEPGGKDYGVPSVKLSFFGRVRRCGMVSPTVFWPIPRVYS GLVRVDRYATSPWPTDDAFRRQVFELVDIAFTQRRKTSRNAFVKWAGSSNESANRLLAASIDPARRGETL SIDDFVRLLRRSDGRDDAAVRSASAS
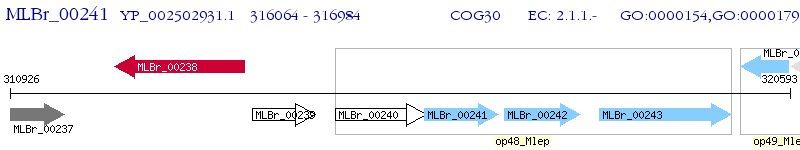
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_00241 | ksgA | - | 100% (306) | dimethyladenosine transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1037 | ksgA | 1e-141 | 85.03% (294) | dimethyladenosine transferase |
| M. gilvum PYR-GCK | Mflv_1933 | ksgA | 1e-128 | 79.58% (284) | dimethyladenosine transferase |
| M. tuberculosis H37Rv | Rv1010 | ksgA | 1e-141 | 85.03% (294) | dimethyladenosine transferase |
| M. abscessus ATCC 19977 | MAB_1131 | ksgA | 1e-114 | 67.12% (292) | dimethyladenosine transferase |
| M. marinum M | MMAR_4478 | ksgA | 1e-116 | 80.38% (260) | dimethyladenosine transferase KsgA |
| M. avium 104 | MAV_1148 | ksgA | 1e-139 | 85.76% (295) | dimethyladenosine transferase |
| M. smegmatis MC2 155 | MSMEG_5438 | ksgA | 1e-119 | 77.49% (271) | dimethyladenosine transferase |
| M. thermoresistible (build 8) | TH_0980 | ksgA | 1e-105 | 75.50% (249) | PROBABLE DIMETHYLADENOSINE TRANSFERASE KSGA |
| M. ulcerans Agy99 | MUL_4650 | ksgA | 1e-115 | 80.00% (260) | dimethyladenosine transferase |
| M. vanbaalenii PYR-1 | Mvan_4800 | ksgA | 1e-129 | 79.23% (284) | dimethyladenosine transferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1933|M.gilvum_PYR-GCK --------------MTIRLLGRTEIRHLAKSIDFRPRKSFGQNFVHDANT
Mvan_4800|M.vanbaalenii_PYR-1 --------------MTIRLLGRTEIRHLAKSIDFRPRKSFGQNFVHDANT
MSMEG_5438|M.smegmatis_MC2_155 ---------------------------MAKEIDFRPRKAFGQNFVHDANT
TH_0980|M.thermoresistible__bu --------------------------------------------------
Mb1037|M.bovis_AF2122/97 ------MCCTSGCALTIRLLGRTEIRRLAKELDFRPRKSLGQNFVHDANT
Rv1010|M.tuberculosis_H37Rv ------MCCTSGCALTIRLLGRTEIRRLAKELDFRPRKSLGQNFVHDANT
MLBr_00241|M.leprae_Br4923 ------MQRKSGCTLTIRLLERTEIRWLVKELECRPRKSLGQNFVHDANT
MMAR_4478|M.marinum_M --------------------------------------------------
MUL_4650|M.ulcerans_Agy99 --------------------------------------------------
MAV_1148|M.avium_104 ---MASVQRKSWCALTIRLLGRTEIRRLAKELEFRPRKSLGQNFVHDANT
MAB_1131|M.abscessus_ATCC_1997 MGRLAGMQRESWCPLTIRLLGRTEIRQLAKDLDFRPTKTLGQNFVHDANT
Mflv_1933|M.gilvum_PYR-GCK VRRIVSASSVNRSDHVLEVGPGLGSLTLALLDRGARVTAVEIDPVLATQL
Mvan_4800|M.vanbaalenii_PYR-1 VRRIVSASSINRSDHVLEVGPGLGSLTLALLDRGAKVTAVEIDPVLANQL
MSMEG_5438|M.smegmatis_MC2_155 VRRIVSASGVHRHDHVLEVGPGLGSLTLALLDRGARVTAVEIDPLLARQL
TH_0980|M.thermoresistible__bu VRRIVSASGINKQDHVLEVGPGLGSLTLGLLDRGAHVTAVEIDSVLARQL
Mb1037|M.bovis_AF2122/97 VRRVVAASGVSRSDLVLEVGPGLGSLTLALLDRGATVTAVEIDPLLASRL
Rv1010|M.tuberculosis_H37Rv VRRVVAASGVSRSDLVLEVGPGLGSLTLALLDRGATVTAVEIDPLLASRL
MLBr_00241|M.leprae_Br4923 VRRVVSTSRVNRSDFVLEVGPGFGSLTLALLDCGAAVSAIEIDPVLAGRL
MMAR_4478|M.marinum_M MRKVVTVSGITRSDQVLEVGPGLGSLTLALLERGAAVTAVEIDPVLAARL
MUL_4650|M.ulcerans_Agy99 MRKVVAVSGTTRSDQVLEVGPGLGSLTLALLERGAAVTAVEIDPVLAARL
MAV_1148|M.avium_104 VRRVVSTSGVSRSDHVLEVGPGLGSLTLALLDRGAHVTAVEIDPVLAERL
MAB_1131|M.abscessus_ATCC_1997 IRRIVSASGVGKGDHVIEVGPGLGSLTLGLLDKGAAVTAVEVDPVLARRL
:*::*:.* : * *:*****:*****.**: ** *:*:*:*.:** :*
Mflv_1933|M.gilvum_PYR-GCK PTTIAAHSHSEVNRLTVLNRDILTFKQSDMTEMPTALVANLPYNVAVPAL
Mvan_4800|M.vanbaalenii_PYR-1 PTTIAAHSHSEVNRLTVLNRDILTFKQSDMTEMPTALVANLPYNVAVPAL
MSMEG_5438|M.smegmatis_MC2_155 PTTIADHSHSEINRLTVLNRDILTLMPGDLEEQPTALVANLPYNVAVPAL
TH_0980|M.thermoresistible__bu PKTIADHSHSEINRLIVLNRDILDLYPQDLETQPTALVANLPYNVAVPAL
Mb1037|M.bovis_AF2122/97 QQTVAEHSHSEVHRLTVVNRDVLALRREDLAAAPTAVVANLPYNVAVPAL
Rv1010|M.tuberculosis_H37Rv QQTVAEHSHSEVHRLTVVNRDVLALRREDLAAAPTAVVANLPYNVAVPAL
MLBr_00241|M.leprae_Br4923 PQTVAEHSNNEIHRLTVCNRDVLSFRRGDLATEPTALVANLPYNVAVPAL
MMAR_4478|M.marinum_M PQTVAEHSDSEIGRLTVVHHDILTLRRQDVADQPTAVVANLPYNVAVPAL
MUL_4650|M.ulcerans_Agy99 PQTVAEHSDSEIGRLTVVHHDILTLRRQDVADQPTAVVANLPYNVAVPAL
MAV_1148|M.avium_104 PHTVAEHSHSEIQRLTVLNRDVLTLRRDELAEPPTAVVANLPYNVAVPAL
MAB_1131|M.abscessus_ATCC_1997 PTTIKEHSHSEFNRFSVIHQDVLNLTPADIQGEPTAMVANLPYNVAVPAL
*: **..*. *: * ::*:* : :: ***:*************
Mflv_1933|M.gilvum_PYR-GCK LHLLAEFPSIRTVMVMVQAEVAERLAAEPGGKDYGVPSAKVRFFGNVRRY
Mvan_4800|M.vanbaalenii_PYR-1 LHLLAEFPSIRTVMVMVQAEVAERLAAEPGGKDYGVPSAKVRFFGNVRRY
MSMEG_5438|M.smegmatis_MC2_155 LHLLAEFPSIRTVMVMVQAEVAERLAAEPGGKDYGVPSAKVRFYGNVRRH
TH_0980|M.thermoresistible__bu LHLLAEFPTIRTVMVMVQAEVAERLAAEPGSKEYGVPSAKVRFYGNVRRY
Mb1037|M.bovis_AF2122/97 LHLLVEFPSIRVVTVMVQAEVAERLAAEPGSKEYGVPSVKLRFFGRVRRC
Rv1010|M.tuberculosis_H37Rv LHLLVEFPSIRVVTVMVQAEVAERLAAEPGSKEYGVPSVKLRFFGRVRRC
MLBr_00241|M.leprae_Br4923 LHLLAEFPSIRTVTVMVQAEVAERLAAEPGGKDYGVPSVKLSFFGRVRRC
MMAR_4478|M.marinum_M LHLLAEFPSIRVVTVMVQAEVAERLAAEPGGKEYGVPSVKVGFYGRVRRC
MUL_4650|M.ulcerans_Agy99 LHLLAEFPSIRVVTVMVQAEVAERLAAEPGGKEYGVPSVKVGFYGRVRRC
MAV_1148|M.avium_104 LHLLAEFPSIRTVTVMVQAEVAERLAAEPGGKEYGVPSVKVRFFGRVRRC
MAB_1131|M.abscessus_ATCC_1997 LRILTEFDSVKTILVMVQSEVADRLTAEPGSRIYGVPSVKVRYFGKVKRL
*::*.** :::.: ****:***:**:****.: *****.*: ::*.*:*
Mflv_1933|M.gilvum_PYR-GCK GMVSPTVFWPIPRVYSGLVRIDRYETSPWPTDTEFQEQVFELIDIAFAQR
Mvan_4800|M.vanbaalenii_PYR-1 GMVSPTVFWPIPRVYSGLVRIDRYETSPWPTDPEFRQQVFELIDVAFAQR
MSMEG_5438|M.smegmatis_MC2_155 GMVSPTVFWPIPRVYSGLVRIDRYETSPWPMDEAFRDEVFNLIDIGFAQR
TH_0980|M.thermoresistible__bu GMVSPTVFWPIPRVYSGLVRLDRYESPKWPMDAEFRERVFELIDIAFAQR
Mb1037|M.bovis_AF2122/97 GMVSPTVFWPIPRVYSGLVRIDRYETSPWPTDDAFRRRVFELVDIAFAQR
Rv1010|M.tuberculosis_H37Rv GMVSPTVFWPIPRVYSGLVRIDRYETSPWPTDDAFRRRVFELVDIAFAQR
MLBr_00241|M.leprae_Br4923 GMVSPTVFWPIPRVYSGLVRVDRYATSPWPTDDAFRRQVFELVDIAFTQR
MMAR_4478|M.marinum_M GMVSPTVFWPIPRVYSGLVRIDRYETTPWPTDENFRRQVFELVDIAFAQR
MUL_4650|M.ulcerans_Agy99 GMVSPTVFWPIPRVYSGLVRIDRHETTPWPTDENFRRQVFELVDIAFAQR
MAV_1148|M.avium_104 GMVSPTVFWPIPRVYSGLVRIDRYPTSPWPTDPAFRRQVFQLVDIAFGQR
MAB_1131|M.abscessus_ATCC_1997 GTVSPAVFWPIPRVYSGLVRIDRHADAAWPSDAGFRKELFDLIDISFAQR
* ***:**************:**: . ** * *: .:*:*:*:.* **
Mflv_1933|M.gilvum_PYR-GCK RKTSRNAFAEWAGSGNESASRLLAASIDPSRRGETLSINDFVRLLQRSAD
Mvan_4800|M.vanbaalenii_PYR-1 RKTSRNAFAEWAGSGNESASRLLAASIDPSRRGETLSINDFVRLLQRSAD
MSMEG_5438|M.smegmatis_MC2_155 RKTSRNAFAEWAGGGNESAERLLAARIDPARRGETLSINDFVRLLQRTKE
TH_0980|M.thermoresistible__bu RKTSRNAFAQWAGSGNESARRLLAASIDPARRGETLSIADFVRLLQR---
Mb1037|M.bovis_AF2122/97 RKTSRNAFVQWAGSGSESANRLLAASIDPARRGETLSIDDFVRLLRR---
Rv1010|M.tuberculosis_H37Rv RKTSRNAFVQWAGSGSESANRLLAASIDPARRGETLSIDDFVRLLRR---
MLBr_00241|M.leprae_Br4923 RKTSRNAFVKWAGSSNESANRLLAASIDPARRGETLSIDDFVRLLRR---
MMAR_4478|M.marinum_M RKTSRNAFLEWAGSGNESANRLLAASIDPARRGETLSIEDFVRLLTRSAD
MUL_4650|M.ulcerans_Agy99 RKTSRNAFLEWAGSGNESANRLLAASIDPARRGETLSIEDFVRLLTRSAD
MAV_1148|M.avium_104 RKTCRNAFVDWAGSGNESADRLLAASIDPARRGETLSIDDFVRLLQRSAD
MAB_1131|M.abscessus_ATCC_1997 RKTSRNAFAEWAGSGDESARRLLAASIDPARRGETLDVADFARLLER---
***.**** .***...*** ***** ***:******.: **.*** *
Mflv_1933|M.gilvum_PYR-GCK WHTVPKADDAGDDADAQAKADGAQVSTL
Mvan_4800|M.vanbaalenii_PYR-1 WHVVPKPESA---ATAVATDEDAQVPTL
MSMEG_5438|M.smegmatis_MC2_155 ALGEDYTKQVARPSASAPSVSAEA----
TH_0980|M.thermoresistible__bu ------TDEVAAPAEK------------
Mb1037|M.bovis_AF2122/97 -SGGSDEATSTGRDARAPDISGHASAS-
Rv1010|M.tuberculosis_H37Rv -SGGSDEATSTGRDARAPDISGHASAS-
MLBr_00241|M.leprae_Br4923 -SDGRDDAAVR-----------SASAS-
MMAR_4478|M.marinum_M TSGGPPDAGVAGDEGQVP---GPVSAT-
MUL_4650|M.ulcerans_Agy99 TSGGPPDAGVAGDEGQVP---GPVSAT-
MAV_1148|M.avium_104 -RGGSDREGTSPPTAGQG-----APAC-
MAB_1131|M.abscessus_ATCC_1997 -------SAIS-----------------