For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LFGEPSWPDHVSMSSPNRAGLFGGLGMTFVGGSVAVSGALAEAPLHTAQALRYAIACLLLLGWVRLTGHP LRRPRGTEWLWLLGVTVSGLVLFNVALVHGSRHAEPAVLAVAVACVPVALATVGPLLEGHRPHTRILVAA VMVSVGAVVVEGLGRADAMGLAWSAVVFVCEAGFTLLAVPVLASHGPAGVSVHTTWLAAVMFGVLALSTE GWRAATRFDAGAVPAIGYLAVGVTAIAFILWYTCVRGLGTGRAGLLTGVAPVAAAVIGIPLTGAVPGVAV WCGVGLIACGLAVGLGVNAERGLVAHIRAKTRTEVDVRGEYPKSRG*
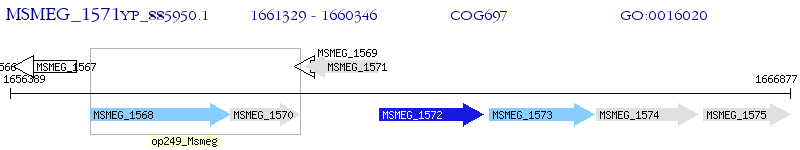
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1571 | - | - | 100% (327) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_2188 | - | 6e-08 | 24.14% (261) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_3276 | - | 3e-07 | 25.17% (294) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3133 | - | 6e-06 | 27.70% (148) | hypothetical protein Mflv_3133 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3762 | - | 4e-09 | 25.56% (270) | hypothetical protein MAB_3762 |
| M. marinum M | MMAR_4903 | - | 3e-06 | 21.26% (301) | hypothetical protein MMAR_4903 |
| M. avium 104 | MAV_4124 | - | 1e-05 | 28.47% (137) | dehydrogenase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5539 | - | 5e-09 | 26.32% (228) | hypothetical protein Mvan_5539 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4903|M.marinum_M -------------------------MPALAMGVTVTLWAFAFVGVR----
Mvan_5539|M.vanbaalenii_PYR-1 ----------------------MSLRGWLLFTAMGVIWGIPYLLIK----
MAB_3762|M.abscessus_ATCC_1997 ----------------------MNDKANFRAGVILALVSAATFGFSGPFA
MSMEG_1571|M.smegmatis_MC2_155 -----------------MLFGEPSWPDHVSMSSPNRAGLFGGLGMTFVGG
Mflv_3133|M.gilvum_PYR-GCK -----------------------MPTDQARTGALMAVAAMLCVQLG-LAV
MAV_4124|M.avium_104 MPDWTAAQLPSFAGRTVIVTGANAGLGEVTARELARVGGHVILAVRNTDK
. .
MMAR_4903|M.marinum_M --SAGRVFS-----------------------------------------
Mvan_5539|M.vanbaalenii_PYR-1 --VAVEGLS-----------------------------------------
MAB_3762|M.abscessus_ATCC_1997 KSLMAAGWS-----------------------------------------
MSMEG_1571|M.smegmatis_MC2_155 SVAVSGALA-----------------------------------------
Mflv_3133|M.gilvum_PYR-GCK AVTLIDDIG-----------------------------------------
MAV_4124|M.avium_104 GRAAADRMAGVATGRVEVRELDLQDLASVRRFADGIDTVDVLVNNAGIMA
.
MMAR_4903|M.marinum_M -------------PGVLALSRLFIAVMVMAVIATIAATYTAYAGRATTRR
Mvan_5539|M.vanbaalenii_PYR-1 -------------VPVLVFARTAVGALVLIPLT-------------LRRG
MAB_3762|M.abscessus_ATCC_1997 -------------LTAVVTARLATGAAVMVVAAC-----------IAAPG
MSMEG_1571|M.smegmatis_MC2_155 -------------EAPLHTAQALRYAIACLLLLG--------WVRLTGHP
Mflv_3133|M.gilvum_PYR-GCK -------------VEGVAWLRLAWAGILFLIIVR---------------P
MAV_4124|M.avium_104 TKHAVTVDGFEGQIGTNHLGHFALTNLLLPKLTDRVVTVSSLMHHFGYIS
:
MMAR_4903|M.marinum_M LRPPSAAALTLVIAYGVAWFGAYSIVISWAEQHIDAGTTALLVNLAPILV
Mvan_5539|M.vanbaalenii_PYR-1 AWAPVLAHWRPVVAFAFFEIIAAWLLLSDAERHITSSLTGLLIAAAPIVA
MAB_3762|M.abscessus_ATCC_1997 WFSEARRHAKVVVLYGIIPVAGAQFCYFGAVRHMSVGVAMLLEFLSPILV
MSMEG_1571|M.smegmatis_MC2_155 LRRPRGTEWLWLLGVTVSGLVLFNVALVHGSRHAEPAVLAVAVACVPVAL
Mflv_3133|M.gilvum_PYR-GCK RRTAFTRNSFLMCVVLGVVTAAITLLFMAALDRIPLGTASALEFLGPLGV
MAV_4124|M.avium_104 LKDLNFRSRPYSAWLAYSQSKLANLLFTSELQRRLDAVPSSLRALAAHPG
. : . .
MMAR_4903|M.marinum_M AVFAGFFLGEG---------------------------------YSARLL
Mvan_5539|M.vanbaalenii_PYR-1 ALLDRLTGGDQP--------------------------------LTVKRL
MAB_3762|M.abscessus_ATCC_1997 IAWTWMITRHR---------------------------------PHTRVL
MSMEG_1571|M.smegmatis_MC2_155 ATVGPLLEGHR---------------------------------PHTRIL
Mflv_3133|M.gilvum_PYR-GCK AVARGRGRGRW---------------------------------------
MAV_4124|M.avium_104 WSHTNLQGNSGRKLGDAAVLAVDRIVSTDADFGARQTLYAVSQDLPGDTF
MMAR_4903|M.marinum_M TGIAIAFAG-----------------------------------------
Mvan_5539|M.vanbaalenii_PYR-1 AGLGTGLAG-----------------------------------------
MAB_3762|M.abscessus_ATCC_1997 AGATLALSG-----------------------------------------
MSMEG_1571|M.smegmatis_MC2_155 VAAVMVSVG-----------------------------------------
Mflv_3133|M.gilvum_PYR-GCK AWPGLAAAG-----------------------------------------
MAV_4124|M.avium_104 VGPRFGLYGRTQPTWRNWPAKRAGTAAALWELSEELTGTSFRSDPDLRRG
. *
MMAR_4903|M.marinum_M -----------------VVLISAGGGGSQTDWLGVGLGLLAAVLYAAGVL
Mvan_5539|M.vanbaalenii_PYR-1 -----------------VAVLAGPELAGGSAWP-VTEVLLVAVCYAIAPL
MAB_3762|M.abscessus_ATCC_1997 -----------------MLLVLNIFSGGRVEPVGIAWGLGAAVCAAYYFI
MSMEG_1571|M.smegmatis_MC2_155 -----------------AVVVEG---LGRADAMGLAWSAVVFVCEAGFTL
Mflv_3133|M.gilvum_PYR-GCK -----------------VLLLTEP-WTGAVDPVGVAFALAAAACWAGYIL
MAV_4124|M.avium_104 RAAGPPSPRRPPGAVTKLVLSATPRGVGGYAMSDANLAAVLALCAALASA
: . *
MMAR_4903|M.marinum_M LQKVA-------LRTVDALSATLWGTLVGFIVTLPFLPAAARELSLASAA
Mvan_5539|M.vanbaalenii_PYR-1 IAARY-------LAEVPAMPLTAACLGLAALIYVG-PAAATWPAEIPSMR
MAB_3762|M.abscessus_ATCC_1997 SSHRAGGK----AEELQPITLATSGLLVGAVAVGGFGLTGIMPLTFSTQD
MSMEG_1571|M.smegmatis_MC2_155 LAVPV-------LASHGPAGVSVHTTWLAAVMFGVLALSTEGWRAATRFD
Mflv_3133|M.gilvum_PYR-GCK LTQKVG------DAVAGINGLAVSMPVAGLVATLTVGPTVIERVTPHILL
MAV_4124|M.avium_104 VGNVVRQRSAQEVTDKPVGHLALFGMLLRDTRWWLGGLGDIGSYVLLAAA
:
MMAR_4903|M.marinum_M DLWW---------LVFLGAGPSAIAFTTWAYALARTNAGV-----TAATT
Mvan_5539|M.vanbaalenii_PYR-1 VLTA---------VALLAVVCTSLAFILFFALIREVGAPR-----ALVIT
MAB_3762|M.abscessus_ATCC_1997 VHVAGWNTPVYVPILVLGVVTTAIAYITGIAAITRLRPSY-----ASLIA
MSMEG_1571|M.smegmatis_MC2_155 AGAVP-------AIGYLAVGVTAIAFILWYTCVRGLGTGR-----AGLLT
Mflv_3133|M.gilvum_PYR-GCK IGLG------------LAVLLPVVPFALELLALRRLTTAA-----FGTLM
MAV_4124|M.avium_104 LDRGS-------VLLVMSLQVTALLFALPIYARMTHHPITGREWAWALLL
:. . : : .
MMAR_4903|M.marinum_M LMVPALVIALSGLLLAEVPNPLGLAGGGACLIGVALARGLL-RLPARSRR
Mvan_5539|M.vanbaalenii_PYR-1 YVNPAVALAAGVIVLNEPLTARHLLGLAMILAGSVLATRRP-VEPMQAAP
MAB_3762|M.abscessus_ATCC_1997 LTEVLCAVIAGWVLLGETIAPIQYAGAAVILAGLMLAHRGS-DTPGEPES
MSMEG_1571|M.smegmatis_MC2_155 GVAPVAAAVIGIPLTGAVPGVAVWCGVGLIACGLAVGLGVN-AERG----
Mflv_3133|M.gilvum_PYR-GCK ALEPAFAMLLGFVVLDQVPGPFGLVGICLVVAAGVGAARTG-ARAAPVPL
MAV_4124|M.avium_104 AVALAVLIAVGDPTGGQQRAPLQTWLVVAAVIGPLLVLGLLGARVWADRP
. .
MMAR_4903|M.marinum_M RTG------TGDQLQGVESLPVGLSDYAASSTAPSGRQL-----------
Mvan_5539|M.vanbaalenii_PYR-1 R-------------------------------------------------
MAB_3762|M.abscessus_ATCC_1997 SGAGLTASLPAGLKSHGDDSELSVTSVCDATTESTPSQ------------
MSMEG_1571|M.smegmatis_MC2_155 ------------LVAHIRAKTRTEVDVRGEYPKSRG--------------
Mflv_3133|M.gilvum_PYR-GCK EVG-----------------------------------------------
MAV_4124|M.avium_104 VAALLLAAVAGSLLAMFAVLMKGVVDILEHNPGQLWRSFELYALVFCGVA
MMAR_4903|M.marinum_M --------------------------------------------------
Mvan_5539|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3762|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_1571|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3133|M.gilvum_PYR-GCK --------------------------------------------------
MAV_4124|M.avium_104 GMIYHQSAYRAGALTASLPTIIVAKPVVGGILGIIVLRETLVAGGWEWVV
MMAR_4903|M.marinum_M -------------------------------------------
Mvan_5539|M.vanbaalenii_PYR-1 -------------------------------------------
MAB_3762|M.abscessus_ATCC_1997 -------------------------------------------
MSMEG_1571|M.smegmatis_MC2_155 -------------------------------------------
Mflv_3133|M.gilvum_PYR-GCK -------------------------------------------
MAV_4124|M.avium_104 LAVTAVVVIVATVGLARGEAASMSAGAGRDVRPTDRPKAASQA