For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ATATSPTHVDSTTSGNQFRLGLLFAVGSALAFGSSGPFAKALIEAGWSPTAAVTARLAGGALLMAIFATF ARPGWVKEALQHSRTVIAYGLIPIAGAQFFYYNAVAHLSVGVALLLEYTAPVLVVGWVWAVTKRRPSAGT FGGVALAVIGIMLVLDVFSGAHINLVGVGFGLAAAVCAACYFMMSNNVDSDGDGLHPYSLAAGGLVVGAV TVAAIGTTGIMPMTFATNPVKLGGFETSWVVPVIALGLIPTALAYTLGIIGIARLKPRFASLVGLSEVMF AVIIAWIMVGEAMTPIQAVGGAVVLLGLALARQGDRSERRAHQITDDVSDDISDEICDACWPETPAHGTA FDETISRG*
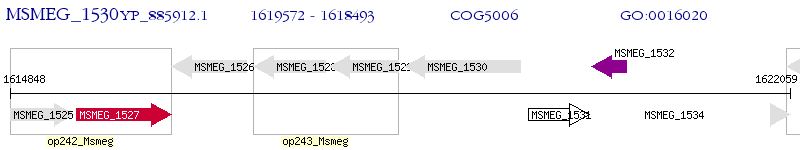
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1530 | - | - | 100% (359) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_3276 | - | 4e-14 | 26.00% (300) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_1505 | - | 2e-10 | 24.15% (294) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4964 | - | 1e-119 | 67.09% (316) | hypothetical protein Mflv_4964 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3762 | - | 5e-89 | 51.18% (297) | hypothetical protein MAB_3762 |
| M. marinum M | MMAR_4903 | - | 1e-14 | 25.63% (277) | hypothetical protein MMAR_4903 |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_0035 | - | 1e-117 | 66.11% (301) | integral membrane protein |
| M. ulcerans Agy99 | MUL_0503 | - | 2e-14 | 25.27% (277) | hypothetical protein MUL_0503 |
| M. vanbaalenii PYR-1 | Mvan_1442 | - | 1e-128 | 67.16% (338) | hypothetical protein Mvan_1442 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4964|M.gilvum_PYR-GCK MAALDSSVTSRTQARADHFRSGLLFALASAFAFGCSGP---FAKALMTAG
Mvan_1442|M.vanbaalenii_PYR-1 MAALDTSRQAR-ADQTHLFRTGLLFAIGSAFAFGSSGP---FAKSLMEAG
MSMEG_1530|M.smegmatis_MC2_155 MATATSPTHVDSTTSGNQFRLGLLFAVGSALAFGSSGP---FAKALIEAG
TH_0035|M.thermoresistible__bu --------LARIHSAVNHFRLGLVFAIASASAFGMSGP---LAKSLLEAG
MAB_3762|M.abscessus_ATCC_1997 ------------MNDKANFRAGVILALVSAATFGFSGP---FAKSLMAAG
MMAR_4903|M.marinum_M ---------------MPALAMGVTVTLWAFAFVGVRSAGRVFSPGVLALS
MUL_0503|M.ulcerans_Agy99 --MASSTTSSQRTVNMPALAMGVTVTLWAFAFVGVRSAGRVFSPGVLALS
: *: .:: : .* .. :: .:: .
Mflv_4964|M.gilvum_PYR-GCK -WSPTAAVTARLAGGALAMAVFATVVRPGWVR-EARHHARTVVLYGIVPI
Mvan_1442|M.vanbaalenii_PYR-1 -WTPTAAVTARLAGGALAMAVFATITYPGWLR-EARRHARTVALYGLVPI
MSMEG_1530|M.smegmatis_MC2_155 -WSPTAAVTARLAGGALLMAIFATFARPGWVK-EALQHSRTVIAYGLIPI
TH_0035|M.thermoresistible__bu -WSPAAAVTARLTGGALVLALFATAIKPGWLR-EAVRHRHTVVLYGIVPV
MAB_3762|M.abscessus_ATCC_1997 -WSLTAVVTARLATGAAVMVVAACIAAPGWFS-EARRHAKVVVLYGIIPV
MMAR_4903|M.marinum_M RLFIAVMVMAVIATIAATYTAYAGRATTRRLRPPSAAALTLVIAYGVAWF
MUL_0503|M.ulcerans_Agy99 RLFIAVMVMAVIATIAATYTAYAGRATTRRLRPPSAAALTLVIAYGVAWF
:. * * :: * . * . . : * **: .
Mflv_4964|M.gilvum_PYR-GCK AGAQLFYFNAVAHLSVGVALLLEYTAPLLVVGWLWATTRHRPSGMTLAGV
Mvan_1442|M.vanbaalenii_PYR-1 AGAQLFYFNAVSHLSVGVALLLEYTAPILVVGWLWATTRRRPTTMTLAGV
MSMEG_1530|M.smegmatis_MC2_155 AGAQFFYYNAVAHLSVGVALLLEYTAPVLVVGWVWAVTKRRPSAGTFGGV
TH_0035|M.thermoresistible__bu AGTQLAYYHAVSHLSVGVALLLEYTAPILVVGWLWLVHRRTPARLTLAGT
MAB_3762|M.abscessus_ATCC_1997 AGAQFCYFGAVRHMSVGVAMLLEFLSPILVIAWTWMITRHRPHTRVLAGA
MMAR_4903|M.marinum_M GAYSIVISWAEQHIDAGTTALLVNLAPILVAVFAGFFLGEGYSARLLTGI
MUL_0503|M.ulcerans_Agy99 GAYSIVISWAEQHIDAGTTALLVNLAPILVAVFAGFFLGEGYSARLLTGI
.. .: * *:..*.: ** :*:** : . : *
Mflv_4964|M.gilvum_PYR-GCK VLAVGGITLVLGIIGPGGVAAAHINLAGVGWGLAAAVCAACYFLMSDKAG
Mvan_1442|M.vanbaalenii_PYR-1 SLAVAGIMLVLGIVGPGGLAGAHINLTGVGWGLAAAVCAACYFLMSDKAG
MSMEG_1530|M.smegmatis_MC2_155 ALAVIGIMLVLDVFS-----GAHINLVGVGFGLAAAVCAACYFMMSNN--
TH_0035|M.thermoresistible__bu ALAVGGMTLVLDVFS-----GAHFDVVGLAWGLTAAVCSASYFLMSDR--
MAB_3762|M.abscessus_ATCC_1997 TLALSGMLLVLNIFS-----GGRVEPVGIAWGLGAAVCAAYYFISSHR--
MMAR_4903|M.marinum_M AIAFAGVVLISAGGG-----GSQTDWLGVGLGLLAAVLYAAGVLLQKV--
MUL_0503|M.ulcerans_Agy99 AIAFAGVVLISAGGG-----GSQTDWLGVGLGLLAAVLYAAGVLLQKV--
:*. *: *: . ..: : *:. ** *** * .: ..
Mflv_4964|M.gilvum_PYR-GCK AMSDKEVADGRPSLHPITLAAAGLAFGAVTVAALGFSGLMPLAFTANDAV
Mvan_1442|M.vanbaalenii_PYR-1 AES---TGDQGP-LHPITLAAGGLIVGAATVAALGATGVMPLTFTANDTV
MSMEG_1530|M.smegmatis_MC2_155 ------VDSDGDGLHPYSLAAGGLVVGAVTVAAIGTTGIMPMTFATNPVK
TH_0035|M.thermoresistible__bu ------VGTGEDGLHPVTLAAGGLLVGAVTVATAGAVGIAPLTFTTGDTV
MAB_3762|M.abscessus_ATCC_1997 ------AGGKAEELQPITLATSGLLVGAVAVGGFGLTGIMPLTFSTQDVH
MMAR_4903|M.marinum_M ---------ALRTVDALSATLWGTLVGFIVTLPF-------LPAAARELS
MUL_0503|M.ulcerans_Agy99 ---------ALRTVDALSATLWGTLVGFIVTLPF-------LPAAARELS
:.. : : * .* .. :. ::
Mflv_4964|M.gilvum_PYR-GCK IAGFTTPWIVPVIALALIPTAIAYTLGIVGIARLRPRFASMVGLSEVMFA
Mvan_1442|M.vanbaalenii_PYR-1 IAGWTTSWIVPVVALALIPTAIAYTLGIVGVARLRPRFASLVGLSEVMFA
MSMEG_1530|M.smegmatis_MC2_155 LGGFETSWVVPVIALGLIPTALAYTLGIIGIARLKPRFASLVGLSEVMFA
TH_0035|M.thermoresistible__bu IAGVTMSWLVPVIGLGVIATAIAYTLGIIGIALLRPRFASLVGLSEVMFA
MAB_3762|M.abscessus_ATCC_1997 VAGWNTPVYVPILVLGVVTTAIAYITGIAAITRLRPSYASLIALTEVLCA
MMAR_4903|M.marinum_M LASAADLWWLVFLGAG--PSAIAFTTWAYALARTNAGVTAATTLMVPALV
MUL_0503|M.ulcerans_Agy99 LASAADLWWLVFLGAG--PSAIAFTTWAYALARTNAGVTAATTLMVPALV
:.. : .: . .:*:*: .:: .. :: * .
Mflv_4964|M.gilvum_PYR-GCK VLAAWMLLGEGMAAAQVIGGAIVLAGLALARQGD----RGE------LVG
Mvan_1442|M.vanbaalenii_PYR-1 VLAAWVLLGEAMTVTQAVGGAVVLAGLALARQGD----RSE------LDE
MSMEG_1530|M.smegmatis_MC2_155 VIIAWIMVGEAMTPIQAVGGAVVLLGLALARQGD----RSERRAHQITDD
TH_0035|M.thermoresistible__bu TVAAWLLIGETVTPVQAVGGVIVLTGLALARQGD----RPVPG----TGD
MAB_3762|M.abscessus_ATCC_1997 VIAGWVLLGETIAPIQYAGAAVILAGLMLAHRGSDTPGEPESSGAGLTAS
MMAR_4903|M.marinum_M IALSGLLLAEVPNPLGLAGGGACLIGVALARGLLRLPARSRRR----TGT
MUL_0503|M.ulcerans_Agy99 IALSGLLLAEVPNPLGLAGGGACLIGVVLARGLLRLPARSRRR----TGT
. :::.* *. * *: **: .
Mflv_4964|M.gilvum_PYR-GCK ETNVGETSVTPTALPQTTPVARGGPATIDP-
Mvan_1442|M.vanbaalenii_PYR-1 AG-----TVTWPDAPAP--------------
MSMEG_1530|M.smegmatis_MC2_155 VSDDISDEICDACWPETPAHGTAFDETISRG
TH_0035|M.thermoresistible__bu TPTTPVDDGPDRRVPELH-------------
MAB_3762|M.abscessus_ATCC_1997 LPAGLKSHGDDSELSVTSVCDATTESTPSQ-
MMAR_4903|M.marinum_M GDQLQGVESLPVGLSDYAASSTAPSGRQL--
MUL_0503|M.ulcerans_Agy99 GDQLQGVESLPVGLSDYAASSTAPSGRQL--
.