For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DATHELAAFLRSRRERLNPQDLGLPMRRAIRRTQGLRREEVAEIAGVSTDYIVRLEQGRGLRPSAEVVEA LAGALRLSPAERAYLYDLARQRPRGADKPATTAAPPLARMVSDLSPLPAMLMNHRFDILAWNPEMAWLLT DFGELPPVQRNAMWVCLMHPEVSAFYVDRDRVVRDAIANLRGAWAAHPEDRVLTDLIGELTSAHNEFARL WAAQDVSGGGRGQKVIRHPEAGEIAVHFEVFMPMQETDLRLMIGRAADEVSQSALDRLVAR*
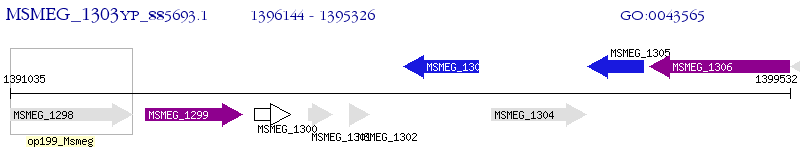
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1303 | - | - | 100% (272) | putative transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_0523 | - | 5e-43 | 40.08% (257) | DNA-binding protein |
| M. smegmatis MC2 155 | MSMEG_1626 | - | 8e-43 | 37.96% (274) | putative DNA-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5170 | - | 2e-30 | 33.47% (248) | XRE family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4641 | - | 5e-32 | 36.86% (255) | putative DNA-binding protein |
| M. marinum M | MMAR_3947 | - | 2e-06 | 33.96% (106) | transcriptional regulatory protein |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0930 | - | 8e-43 | 37.22% (266) | helix-turn-helix domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5170|M.gilvum_PYR-GCK MDTRAQ----------------------RRLALGEFLRARREAIRRADVG
MAB_4641|M.abscessus_ATCC_1997 MAGTDE----------------------RRRELGAFLRVRREHLVRADYG
MSMEG_1303|M.smegmatis_MC2_155 MDAT--------------------------HELAAFLRSRRERLNPQDLG
Mvan_0930|M.vanbaalenii_PYR-1 MQCVGALRLDGALALSDHGRTASSDDRARREALAEFLRARRESLQPEDIG
MMAR_3947|M.marinum_M -----------------------------MPNRGGFVHAARERA------
. *:: **
Mflv_5170|M.gilvum_PYR-GCK LP-ELPRARTGGLRREEVAVTAGVSVTWYTWLEQGRDINPSKQVLDAVAD
MAB_4641|M.abscessus_ATCC_1997 LP-PVGRSRTTGLRREEIAYLSGVSVTWYTWLEQGRDINPSRQVLEAIAL
MSMEG_1303|M.smegmatis_MC2_155 LPMRRAIRRTQGLRREEVAEIAGVSTDYIVRLEQGRGLRPSAEVVEALAG
Mvan_0930|M.vanbaalenii_PYR-1 IP-RRGRRRVKGLRRHEVADAAAVSVTWYTWLEQGRDIHATPQVIEALAR
MMAR_3947|M.marinum_M -----------GLTCRELAEKAGCSLRYVHDIEQG--CSPSIRVLNGLVR
** .*:* :. * : :*** .: .*::.:.
Mflv_5170|M.gilvum_PYR-GCK ALRLSPAEHEYVLALGGYA---VDAAVSRGPLPPHIQHFLDALEGYPAFA
MAB_4641|M.abscessus_ATCC_1997 EMRLSAAERTYLLELAGFAPLPHSATPDPAPVPDHLLRLVENLVPSPAFV
MSMEG_1303|M.smegmatis_MC2_155 ALRLSPAERAYLYDLARQR--PRGADKPATTAAPPLARMVSDLSPLPAML
Mvan_0930|M.vanbaalenii_PYR-1 ALQLDDSEHSYLCRLAGVT--PKATQLGRTQAGDSLIALVDSLLPNPAQL
MMAR_3947|M.marinum_M ALQLSAWEQRFLFAACGRI-----LPDPALTDTDISQRYLDALQPHPAAA
::*. *: :: :. * **
Mflv_5170|M.gilvum_PYR-GCK ITPDWGIAAWNTTYAALYPNVERVPETDRNLLWLVFTDPYIHELLPNWDT
MAB_4641|M.abscessus_ATCC_1997 VSADWSIGGWNRAYELLYPGIASIVPADRNLLRFIFTDPYVRQMLPDWPI
MSMEG_1303|M.smegmatis_MC2_155 MNHRFDILAWNPEMAWLLTDFGELPPVQRNAMWVCLMHPEVSAFYVDRDR
Mvan_0930|M.vanbaalenii_PYR-1 MLPATDLINWNDAYSRLFVDPSTLAAEDRNALWIQVMCPEVRHRLVDWEA
MMAR_3947|M.marinum_M LHPSWRFKAWNQQWQRLFQGVLLAPSMHQ----WLYLSTTSRRILRNWDE
: : ** * . .: . :
Mflv_5170|M.gilvum_PYR-GCK DSRRFLAEFRADAGPRVGEPSYTALVSRLSSESPHFAAAWQATSIERFTS
MAB_4641|M.abscessus_ATCC_1997 TSRRFLAEYRAEAGALLGDPRHRALVDRLQQQSPEFARAWAEHEVGRFTS
MSMEG_1303|M.smegmatis_MC2_155 VVRDAIANLRGAWAAHPEDRVLTDLIGELTSAHNEFARLWAAQDVSGGGR
Mvan_0930|M.vanbaalenii_PYR-1 ETQLVVGRFRAEAAKYPGDPHFRRIIDRLSAESTLFRSIWARHEVQGFED
MMAR_3947|M.marinum_M VSQWCVGWLR--YGLAVDVDAVKPMVSALMP-VTEFRREWEAQVIPVDPA
: :. * . ::. * * * :
Mflv_5170|M.gilvum_PYR-GCK RERLFRHPAAGELRLEHHQVAPVDVPDIQIVAYLPVPDSVASERLRMLTG
MAB_4641|M.abscessus_ATCC_1997 RERRFHHPEAGELIFEHHRLSPSDLPDLHIVIYLPLDGTNTRTKLEALLS
MSMEG_1303|M.smegmatis_MC2_155 GQKVIRHPEAGEIAVHFEVFMPMQETDLRLMIGR-AADEVSQSALDRLVA
Mvan_0930|M.vanbaalenii_PYR-1 RVKTIEHPEVGEVRVRLVKLRPLDHPHLLLLVHM-LDDDESRARIGRLLS
MMAR_3947|M.marinum_M TRLWVVRDGGTELRIDMRFWHASPMPGGLLLGVILDAG------------
. : *: . . . :: .
Mflv_5170|M.gilvum_PYR-GCK QADTAR
MAB_4641|M.abscessus_ATCC_1997 GGR---
MSMEG_1303|M.smegmatis_MC2_155 R-----
Mvan_0930|M.vanbaalenii_PYR-1 R-----
MMAR_3947|M.marinum_M ------