For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PRFMVHGAFTLVTVAGMDLNTVEVVDRPTRREEVWPLGAGDAVLAGGTWLFSEPQPQLRRLVDLTGLGWP PVTLSDSGIELAATCTLAEVSRLSADLQEHRTQWTAAPLLHQCCGALLASFKIWHCATVGGNICLSFPAG AMISLCTALDGEATVWRADGNDFRLPVAALVTGAGTNLLAAADVLRSVHLPAAALRARTSYRKLAPSPLG RSGIVVIGRRDTVPDGGRFVLSVTAATVRPFVFTFPGVPDQDQLTDALGAIPPDAWSDDPHGDPDWRRAM TLVLAEQVRSELT*
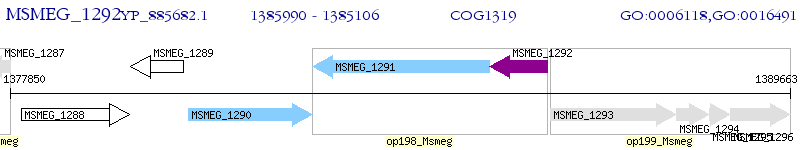
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1292 | - | - | 100% (294) | FAD binding domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_0744 | - | 3e-05 | 22.97% (222) | carbon monoxide dehydrogenase medium chain |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2926c | - | 3e-66 | 48.74% (277) | putative oxidoreductase (molybdopterin dehydrogenase) |
| M. marinum M | MMAR_4946 | - | 1e-105 | 66.67% (276) | hypothetical protein MMAR_4946 |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_0174 | - | 6e-05 | 27.61% (163) | PROBABLE CARBON MONOXYDE DEHYDROGENASE (MEDIUM CHAIN) |
| M. ulcerans Agy99 | MUL_0458 | - | 2e-99 | 66.04% (265) | hypothetical protein MUL_0458 |
| M. vanbaalenii PYR-1 | Mvan_5280 | - | 1e-104 | 66.43% (277) | molybdopterin dehydrogenase, FAD-binding |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4946|M.marinum_M -----------------MDLN---------------SVEIVNTP------
MUL_0458|M.ulcerans_Agy99 -MRHWPPGSTGCASIP-ANISPRAAPPGSTPRSSSPSTSLTSTPNTTCRC
Mvan_5280|M.vanbaalenii_PYR-1 -----------------MDLN---------------TVEAVHIP------
MSMEG_1292|M.smegmatis_MC2_155 MPRFMVHGAFTLVTVAGMDLN---------------TVEVVDRP------
MAB_2926c|M.abscessus_ATCC_199 -----------------MDLY---------------SIDSVLIP------
TH_0174|M.thermoresistible__bu -----------------MQVPG--------------PFEYERATS-----
:: . . .
MMAR_4946|M.marinum_M -------------------------SRRDELWPLEPTDAILAGGTWLFSE
MUL_0458|M.ulcerans_Agy99 CCRRTRIRHTGGVNGSQLRRNRQYPSRRDELWPLEPTDAILAGGTWLFSE
Mvan_5280|M.vanbaalenii_PYR-1 -------------------------TRRDEVWPVGPSDAILAGGTWLFSE
MSMEG_1292|M.smegmatis_MC2_155 -------------------------TRREEVWPLGAGDAVLAGGTWLFSE
MAB_2926c|M.abscessus_ATCC_199 -------------------------RDRADLSGLGTQDGVLAGGTWLMSQ
TH_0174|M.thermoresistible__bu -----------------------VDHAIGLLDRLGDGAMIVAGGHSLLPM
: : ::*** *:.
MMAR_4946|M.marinum_M PNSHIRRLVDITGLGWQPIAFGDNGIDLAATCTLAEISTLSGRLP---AE
MUL_0458|M.ulcerans_Agy99 PNSHIRRLVDRTGLGWQPIAFGDDGIDLAATCTLAEISTLSGRLP---AD
Mvan_5280|M.vanbaalenii_PYR-1 PQPQVSRLIDITRLGWPPITFSDDGMEIAATCSLAEIGSLTNTLPSDRPD
MSMEG_1292|M.smegmatis_MC2_155 PQPQLRRLVDLTGLGWPPVTLSDSGIELAATCTLAEVSRLSADLQEHRTQ
MAB_2926c|M.abscessus_ATCC_199 PQPGLRRLVDLTGMRWPDLVVDAGGLEVAATCSIERL--INASYP---PA
TH_0174|M.thermoresistible__bu MKLRIANPEYLVDINDLAGELG-YIVVNADAVRIGAMTRHREALES--AE
: : . . : .. : * : : : .
MMAR_4946|M.marinum_M WTAGALLYQCCTAMLASAKIWRTATIGGNVCLSFPAGAMISLLSALDARV
MUL_0458|M.ulcerans_Agy99 WTAGAVLYQCCTAMLASAKIWRTATIGANVCLSFPAGAMISLLSALDARV
Mvan_5280|M.vanbaalenii_PYR-1 WPAAPLFGQCCAALLASFKILGSATVGGNICLSFPAGAMISLASALDGTV
MSMEG_1292|M.smegmatis_MC2_155 WTAAPLLHQCCGALLASFKIWHCATVGGNICLSFPAGAMISLCTALDGEA
MAB_2926c|M.abscessus_ATCC_199 WQATALFGQCARALLASFKIWRTATVGGNICLGLPAGAMISLCAALDAEL
TH_0174|M.thermoresistible__bu LAQCCPIFADAERVIADPVVRNRGTVGGSLCQADPAEDLSTVCTVLDAVC
: . ::*. : .*:*..:* . ** : :: :.**.
MMAR_4946|M.marinum_M TVWCADGNDYTLPVAEFVTGQAANVLVPGDVLRSFHLPAAALRARTAFRK
MUL_0458|M.ulcerans_Agy99 TVWCADGNDYTLPVAEFVTGQAVNVLVPGDVLRSFHLPAAALRARTAFRK
Mvan_5280|M.vanbaalenii_PYR-1 TVWSADGSDYVVPIGEFVTGAGSNVLRSGDVLRSLWLPAHALRSPTAFRK
MSMEG_1292|M.smegmatis_MC2_155 TVWRADGNDFRLPVAALVTGAGTNLLAAADVLRSVHLPAAALRARTSYRK
MAB_2926c|M.abscessus_ATCC_199 HIWRHDGTDERCTITEFVTGINETVLHPGDVLRAITFPSSALSQPTALRK
TH_0174|M.thermoresistible__bu VVRGPAGR-REIGIDDFLVGPYETALAHNEILLEVKIPLRPNSS-NAYAK
: * : ::.* . * ::* . :* . .: *
MMAR_4946|M.marinum_M LAP------SALGRSGIVVIGRRGAESDGGGFVLSITAATVRPFVFEFPT
MUL_0458|M.ulcerans_Agy99 LAP------SALGRSGIVVIGRRGAESDGGGFVLSITAATVRPFVFEFPT
Mvan_5280|M.vanbaalenii_PYR-1 LAP------SPLGRSGVVVVGRR--DEDDGQFVLSVTAATVRPHVFRFPL
MSMEG_1292|M.smegmatis_MC2_155 LAP------SPLGRSGIVVIGRRDTVPDGGRFVLSVTAATVRPFVFTFPG
MAB_2926c|M.abscessus_ATCC_199 IAY------SPLGRSGALVTGRL----DGADIVLGITAATRRPIVLRVP-
TH_0174|M.thermoresistible__bu VERRVGDWAVTAAGAAVTLDGQTVTAARIGLTAVNPDAAALAEISRRLTG
: . . :. : *: . .:. **: ..
MMAR_4946|M.marinum_M LPGTDELRAAHAGIPDGAWT--RDAHGDPDWRAAVSLVLAEQILAELA--
MUL_0458|M.ulcerans_Agy99 LPGTDELRAAHAGIPDGAWT--RDAHGDPDWRAAVSLVLAEQILAELA--
Mvan_5280|M.vanbaalenii_PYR-1 PPTAGEIRTALAAIAETEFT--RDAHGDPDWRRAVTLTLAEQIRRELT--
MSMEG_1292|M.smegmatis_MC2_155 VPDQDQLTDALGAIPPDAWS--DDPHGDPDWRRAMTLVLAEQVRSELT--
MAB_2926c|M.abscessus_ATCC_199 -AEARRAREAVAAIPGVEFH--TDAHGTAEWRGAVTTQLAGEVAAELTGA
TH_0174|M.thermoresistible__bu RPATEETFAEAGRLAAQACEPVTDVRGTADYKRHLAAELTTRTLRTAVQR
. . . :. * :* .::: :: *: . .
MMAR_4946|M.marinum_M ------------
MUL_0458|M.ulcerans_Agy99 ------------
Mvan_5280|M.vanbaalenii_PYR-1 ------------
MSMEG_1292|M.smegmatis_MC2_155 ------------
MAB_2926c|M.abscessus_ATCC_199 LS----------
TH_0174|M.thermoresistible__bu IQQRHGGAQPCR