For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGLVQALVGGILIGGLYVAISIGFSLSFGVLDVVDLAVGMWVVIGAFAAIVVSEATGIDAFALLPAVFVV FGIVGWIIAPLIYRVRMSKYALPALMGLAFTFGLATLIRGGLLTVFGYNPRTVPTSLFTGNVSVLGITAP MIRVAGFGFAVIATALFLAFLFYTRTGLAIRATAQSKENAGLMGIDVRRISSLVYAIYTGLTAMAGALLG AIYAMTPEVGLRYTLFAFFVVVLAGLGSVVGVMVAGLFLGILQSLVTTYVGANYTLLVVFAVLFVALLLF PQGISRRGIA*
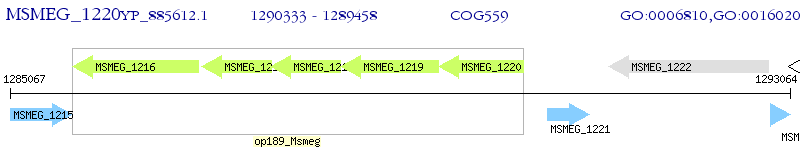
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1220 | - | - | 100% (291) | ABC-type transport system permease protein I |
| M. smegmatis MC2 155 | MSMEG_2981 | - | 1e-31 | 30.04% (283) | branched-chain amino acid ABC-type transport system, permease |
| M. smegmatis MC2 155 | MSMEG_6879 | - | 1e-27 | 27.76% (281) | integral membrane protein of the ABC-type Nat permease for |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3799 | - | 1e-30 | 28.27% (283) | inner-membrane translocator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00397 | - | 3e-09 | 22.70% (282) | putative transporter protein |
| M. abscessus ATCC 19977 | MAB_2625c | - | 3e-18 | 26.09% (322) | branched chain amino acid ABC transporter |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_2329 | - | 9e-33 | 30.74% (283) | branched-chain amino acid ABC-type transport system, permease |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2595 | - | 1e-30 | 29.02% (286) | inner-membrane translocator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3799|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2595|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
MSMEG_1220|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2625c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 MDHTTKCDAEQYFQAIVTSMADGVIVVDIDGRIESINPAATRILGLRAHD
Mflv_3799|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2595|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
MSMEG_1220|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2625c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 VVDMKHGHPFCFYDTDNQRVDLEREVMRVVRREVTTVSKVVGIDQHSGQR
Mflv_3799|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2595|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
MSMEG_1220|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2625c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 LWLSVNVSLLAYKAPPHSALVVSFSDISAHHLSIERLTYEATHDCLTGLA
Mflv_3799|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2595|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
MSMEG_1220|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2625c|M.abscessus_ATCC_199 ------------------------------------------------MI
MLBr_00397|M.leprae_Br4923 NRRFAEDQITKSLQHDERSRLAAVLLLDLDDFKVINDSLGHDVGDAVLQT
Mflv_3799|M.gilvum_PYR-GCK -----------------------------------MDVVIGQLATGLSLG
Mvan_2595|M.vanbaalenii_PYR-1 -----------------------------------MDVVIGQLATGLSLG
TH_2329|M.thermoresistible__bu -----------------------------------MDILIGQLATGLSLG
MSMEG_1220|M.smegmatis_MC2_155 -----------------------------------MTGLVQALVGGILIG
MAB_2625c|M.abscessus_ATCC_199 SGWMAQTGVQASTISFDLHG--------------LWSSIGQLTIDGLAWG
MLBr_00397|M.leprae_Br4923 VAQRLRSAVRPDDVVARLGGDEFIVLLRGPLSDMNANDVAKRLHTTLSES
: : .
Mflv_3799|M.gilvum_PYR-GCK SILLLAALGLSLTFGQMGVIN--MAHGEFIMAGCYTAYVVQQVVS-----
Mvan_2595|M.vanbaalenii_PYR-1 SILLLAALGLALTFGQMGVIN--MAHGEFIMAGCYTAYVVQQLVS-----
TH_2329|M.thermoresistible__bu SILLLAALGLALTFGQMGVIN--MAHGEFIMVGCYTAYVVQQIVS-----
MSMEG_1220|M.smegmatis_MC2_155 GLYVAISIGFSLSFGVLDVVD--LAVGMWVVIGAFAAIVVSEATG-----
MAB_2625c|M.abscessus_ATCC_199 AIYALVAVGYTLVFGVLRLIN--FAHSEVFMLGMFGAYFALDVVLGFSPG
MLBr_00397|M.leprae_Br4923 LVVDQLTVPIGASVGILEVRPDDRRRAADILRDADSAMYAAKNKKQ----
: :: .* : : . .: . * . .
Mflv_3799|M.gilvum_PYR-GCK ----NAGVSLILSLVIGF-LVGGAMGALLEV-------TLIQRMYHRPLD
Mvan_2595|M.vanbaalenii_PYR-1 ----SAGVSLIISLVVGF-FIGGAMGALLEV-------TLIQRMYDRPLD
TH_2329|M.thermoresistible__bu ----DAGVSLLVSLFVGF-LVGGAMGALLEV-------TLIRRMYHRPLD
MSMEG_1220|M.smegmatis_MC2_155 ----IDAFALLPAVFVVFGIVGWIIAPLIYR-------VRMSKYALPALM
MAB_2625c|M.abscessus_ATCC_199 GNAYNKGVSLTVLYLGVAMLVAMAVSATTALGLEAVAYRPLRRRGARPLT
MLBr_00397|M.leprae_Br4923 CAVTPQQLVPFVALIALFVFFTAAAGAKFYAP------SNLLVILQQTVV
. . :. .. : .:
Mflv_3799|M.gilvum_PYR-GCK TLLVTFGVGLILQQ-----VARDIFG-------------------APAVN
Mvan_2595|M.vanbaalenii_PYR-1 TLLVTFGVGLILQQ-----VARDIFG-------------------APAVN
TH_2329|M.thermoresistible__bu TLLVTFGVGLILQQ-----VARDIFG-------------------APAVN
MSMEG_1220|M.smegmatis_MC2_155 GLAFTFGLATLIRG-----GLLTVFG-------------------YNPR-
MAB_2625c|M.abscessus_ATCC_199 FLITAIGMSFVIQE-----FVHWVLPKIAKGYGGSNAQQPLMLVTPRPVW
MLBr_00397|M.leprae_Br4923 LAIVGYGMTFVIMAGSVDLSVGSIVALTGVTAALVAAQNQFAAIVTALLV
*: :: :.
Mflv_3799|M.gilvum_PYR-GCK VIAPVWLSGGVEIFGAVVPKTRIF--------------------------
Mvan_2595|M.vanbaalenii_PYR-1 VVAPEWLSGGVEILGAVVPKTRIF--------------------------
TH_2329|M.thermoresistible__bu VLAPAWLGGGVEIFGAVVPKTRLF--------------------------
MSMEG_1220|M.smegmatis_MC2_155 TVPTSLFTGNVSVLGITAPMIRVA--------------------------
MAB_2625c|M.abscessus_ATCC_199 TFGPFDVSGHHIPFQATITNVTVV--------------------------
MLBr_00397|M.leprae_Br4923 GLAAGMVNGIVFAYGKIPSFVSTLGMLQVCRGITLMISDSSAKPMPFHGI
. . . * .
Mflv_3799|M.gilvum_PYR-GCK ---------ILVLAIACVAVLAAVLKVSPMGRRIRAVVQNRDLAETSGIS
Mvan_2595|M.vanbaalenii_PYR-1 ---------ILVLAVVCVAVLAVVLKISPMGRRIRAVVQNRDLAETSGIS
TH_2329|M.thermoresistible__bu ---------ILVLAVVCVAALAAVLKYSPAGRRIRAVVQNRDLAETSGIS
MSMEG_1220|M.smegmatis_MC2_155 ---------GFGFAVIATALFLAFLFYTRTGLAIRATAQSKENAGLMGID
MAB_2625c|M.abscessus_ATCC_199 ---------VVAAALVLAFVTDIAINHTKFGRGIRAVAQDSTTATLMGVS
MLBr_00397|M.leprae_Br4923 LGAMGAMPWILIVCLFVTILAGILFQFTMFGRWVKAIGGNERVATLAGVP
. .: . : : * ::* . * *:
Mflv_3799|M.gilvum_PYR-GCK SRKTDITTFFIGSGLAAVAGVALTLIG--STSPTIGQSYLIDAFLVVVIG
Mvan_2595|M.vanbaalenii_PYR-1 SRRTDITTFFIGSGLASVAGVALTLIG--STSPTIGQSYLIDAFLVVVVG
TH_2329|M.thermoresistible__bu SRTTDVTTFFIGSGLAAVAGVALTLIG--STSPTIGQSYLIDAFLVVVVG
MSMEG_1220|M.smegmatis_MC2_155 VRRISSLVYAIYTGLTAMAGALLGAIY--AMTPEVGLRYTLFAFFVVVLA
MAB_2625c|M.abscessus_ATCC_199 RERIIMATFVIGGLLAGAAALLYTLKVPQGIIYSGGFLLGIKAFSAAVLG
MLBr_00397|M.leprae_Br4923 TRGIKVAIFAICGLTAGLGGIVLASRLG-AGTPTAATGFEIDVIAAVVIG
. : * :. .. . . : .: ..*:.
Mflv_3799|M.gilvum_PYR-GCK ------GLGQIKGTVIAAFALGFLNSFIEYNTTASLAKVIVFVIIVIVLQ
Mvan_2595|M.vanbaalenii_PYR-1 ------GLGQIKGTVIAAFTLGFLNSFIEYNTTASLAKVIVFVIIVIFLQ
TH_2329|M.thermoresistible__bu ------GLGQIKGTVIAALGLGFLNSFIEYSTTASLAKVIVFALIVIFLQ
MSMEG_1220|M.smegmatis_MC2_155 ------GLGSVVGVMVAGLFLGILQSLVTTYVGANYTLLVVFAVLFVALL
MAB_2625c|M.abscessus_ATCC_199 ------GIGNLRGALLGGLILGVMENYGQAVFGTQWRDVVAFVLLVLVLM
MLBr_00397|M.leprae_Br4923 GTPLTGGLGRLSGTLIGAIIISMLSNGMVFMGVGNAASQIIKGIMLAAAV
*:* : *.::..: :..:.. . : ::.
Mflv_3799|M.gilvum_PYR-GCK VRPQGLFTVRTRSLV-
Mvan_2595|M.vanbaalenii_PYR-1 ARPQGLFTVRTRSLV-
TH_2329|M.thermoresistible__bu VRPQGLFTVRTRSLA-
MSMEG_1220|M.smegmatis_MC2_155 LFPQGISRRGIA----
MAB_2625c|M.abscessus_ATCC_199 FRPTGILGESLGRAKA
MLBr_00397|M.leprae_Br4923 FVFLQRRKIGIIK---