For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLAVAASLLLVITCTQDVESRGEPAGGPPAIATDLDVPWGIAFLPDGSALIAERDSAAIKHLTAPGVVEQ IGAVPGVAPRGEGGLLGLATAGANVYAYLTTGGDNRVVQMRFADGALTNPTPILTGIPAGSRHDGGRIAF GPDGKLYVATGEIANPPLAQDRTSLAGKILRVNPDGSVPADNPFPGSPVWSYGHRNIQGLAWDAAGRLWA TEYGADRVDELNLIRPGANYGWPEAEGRGGIEGMVDPVVEWGTGEASPSGLAFYGGSLWVAALRGERLFQ IPVAGDGTVGTPVPLFVGQYGRLRTVVAAPDGSLWFTTSNRDGRGSVRAGDDRILQFRP
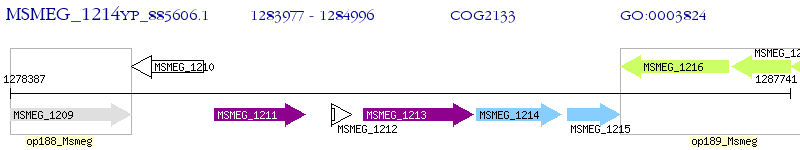
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1214 | - | - | 100% (339) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_2369 | - | 1e-18 | 29.37% (303) | LppZ protein |
| M. smegmatis MC2 155 | MSMEG_0214 | - | 3e-07 | 22.83% (381) | L-sorbosone dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3031 | lppZ | 2e-17 | 28.20% (305) | lipoprotein LppZ |
| M. gilvum PYR-GCK | Mflv_5245 | - | 1e-27 | 31.62% (272) | glucose/sorbosone dehydrogenases-like protein |
| M. tuberculosis H37Rv | Rv3006 | lppZ | 3e-17 | 28.20% (305) | lipoprotein LppZ |
| M. leprae Br4923 | MLBr_01699 | - | 4e-17 | 27.48% (302) | putative lipoprotein |
| M. abscessus ATCC 19977 | MAB_3333 | - | 3e-17 | 29.53% (298) | lipoprotein LppZ |
| M. marinum M | MMAR_1706 | lppZ | 2e-17 | 28.62% (311) | lipoprotein LppZ |
| M. avium 104 | MAV_3855 | - | 6e-19 | 28.80% (316) | LppZ protein |
| M. thermoresistible (build 8) | TH_2267 | - | 1e-137 | 68.97% (348) | oxidoreductase |
| M. ulcerans Agy99 | MUL_1943 | lppZ | 5e-18 | 28.62% (311) | lipoprotein LppZ |
| M. vanbaalenii PYR-1 | Mvan_1010 | - | 6e-32 | 31.68% (303) | PKD domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3031|M.bovis_AF2122/97 --------------------------------------------------
Rv3006|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1706|M.marinum_M --------------------------------------------------
MUL_1943|M.ulcerans_Agy99 --------------------------------------------------
MAV_3855|M.avium_104 --------------------------------------------------
MLBr_01699|M.leprae_Br4923 --------------------------------------------------
MAB_3333|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_1214|M.smegmatis_MC2_155 --------------------------------------------------
TH_2267|M.thermoresistible__bu --------------------------------------------------
Mflv_5245|M.gilvum_PYR-GCK MGVSADTVRTGRPPGGHSRYIGRVGGLAFALGIGAAMLTGTAVASAETSE
Mvan_1010|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3031|M.bovis_AF2122/97 --------------------------------------------------
Rv3006|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1706|M.marinum_M --------------------------------------------------
MUL_1943|M.ulcerans_Agy99 --------------------------------------------------
MAV_3855|M.avium_104 --------------------------------------------------
MLBr_01699|M.leprae_Br4923 --------------------------------------------------
MAB_3333|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_1214|M.smegmatis_MC2_155 --------------------------------------------------
TH_2267|M.thermoresistible__bu --------------------------------------------------
Mflv_5245|M.gilvum_PYR-GCK PSSSSAESSAGPSATERESSKPTVESRSESDADADAESDASEDSDDADAP
Mvan_1010|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3031|M.bovis_AF2122/97 --------------------------------------------------
Rv3006|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1706|M.marinum_M --------------------------------------------------
MUL_1943|M.ulcerans_Agy99 --------------------------------------------------
MAV_3855|M.avium_104 --------------------------------------------------
MLBr_01699|M.leprae_Br4923 --------------------------------------------------
MAB_3333|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_1214|M.smegmatis_MC2_155 --------------------------------------------------
TH_2267|M.thermoresistible__bu --------------------------------------------------
Mflv_5245|M.gilvum_PYR-GCK DESTEGAGDDVDDRASRNTTQPTEVEEPATGTSRAQAEAEEPSFFERIFD
Mvan_1010|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3031|M.bovis_AF2122/97 --------------------------------------------------
Rv3006|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1706|M.marinum_M --------------------------------------------------
MUL_1943|M.ulcerans_Agy99 --------------------------------------MAAGGGAADNVR
MAV_3855|M.avium_104 --------------------------------------------------
MLBr_01699|M.leprae_Br4923 --------------------------------------------------
MAB_3333|M.abscessus_ATCC_1997 ----------------------------------------MLIRSSPATG
MSMEG_1214|M.smegmatis_MC2_155 --------------------------------------------------
TH_2267|M.thermoresistible__bu --------------------------------------------------
Mflv_5245|M.gilvum_PYR-GCK NKTPTISHNPADDTVIEGRVIGNLHPEDPDSTQLTYTATRPASGTVTIES
Mvan_1010|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3031|M.bovis_AF2122/97 ----MWTTRLVRSGLAALCAAVLVSSGCARFNDAQSQPFTTEPELRPQPS
Rv3006|M.tuberculosis_H37Rv ----MWTTRLVRSGLAALCAAVLVSSGCARFNDAQSQPFTTEPELRPQPS
MMAR_1706|M.marinum_M ----MRIRRSIRYALAALCAVTLVSSGCARFNDAQSQPFTTNPELRPQPS
MUL_1943|M.ulcerans_Agy99 SLSVMRIRRSIRYALAALCAVTLVSSGCARFNDAQSQPFTTNPELRPQPS
MAV_3855|M.avium_104 ---------------------MLLTSGCARFNDAQSQPFTTAPELKPQPS
MLBr_01699|M.leprae_Br4923 ----MRIGRSVRSGFTALCAALLILSSCARFNNTQSQPFTTKPELRPQPS
MAB_3333|M.abscessus_ATCC_1997 PTNEPSGRKLLVGSMAALAAAVTLLSGCVRFDPTQDQPFTEVPRRGVAQT
MSMEG_1214|M.smegmatis_MC2_155 ---------------------------------MLAVAASLLLVITCT-Q
TH_2267|M.thermoresistible__bu --------------------VAVGRIRRTTVRAAVALMCLALLAAACDGE
Mflv_5245|M.gilvum_PYR-GCK DGTFTYTPGATYTGQDRFRVTVSDARSGFHIHGAAGLLNLFTFGLLGSSG
Mvan_1010|M.vanbaalenii_PYR-1 --------------------------------------------MLGSSG
Mb3031|M.bovis_AF2122/97 STPPPPPPLPPVPFPKECPAPGVMQGCLESTSGLIMGIDSKTALVAERIT
Rv3006|M.tuberculosis_H37Rv STPPPPPPLPPVPFPKECPAPGVMQGCLESTSGLIMGIDSKTALVAERIT
MMAR_1706|M.marinum_M STPPPPPPLPPTPFPKECPAPGVMQGCLDSTSGLIMGLDSKSALVAERTT
MUL_1943|M.ulcerans_Agy99 STPPPPPPLPPTPFPKECPAPGVMQGCLDSTSGLIMGLDSKSALVAERTT
MAV_3855|M.avium_104 STPPPPPPLPPTPFPKACPAPGVMQGCLESTSGLIMGPDSKTALVAERTT
MLBr_01699|M.leprae_Br4923 SIPPPPP-LPLTPFPKQCPAPGVMQGCLESTSGLIMGIDNKTALVAERTT
MAB_3333|M.abscessus_ATCC_1997 TTTPPPP-LPGKPFPKQCPAEGVMQGCLEATSGLIMGSDGKTALVAERAT
MSMEG_1214|M.smegmatis_MC2_155 DVESRGEPAGGPPA---------IATDLDVPWGIAFLPDGS-ALIAERDS
TH_2267|M.thermoresistible__bu AVPAPPAPPEQPPAQTGESGARDVVTDLGVPWGIAFLPDGS-ALIAERDS
Mflv_5245|M.gilvum_PYR-GCK HRSTQDVFIGSQRAT--------VVSGLNSPVDFRFLPDGR-ILVAEKAG
Mvan_1010|M.vanbaalenii_PYR-1 HRSTQTVFIGYERAT--------VVSGLNSPVDFRFLPDGR-ILVAEKAG
. : * . .: : *. *:**:
Mb3031|M.bovis_AF2122/97 GAVEEISIS-AEPKVKTVIPVDPAGDGGLMDIVLSPTYSQDR--LMYAYI
Rv3006|M.tuberculosis_H37Rv GAVEEISIS-AEPKVKTVIPVDPAGDGGLMDIVLSPTYSQDR--LMYAYI
MMAR_1706|M.marinum_M GAVKEVSVS-AEPKVKVVIPVDPAGDGGLMDIVLSPTYSQDR--LMYAYI
MUL_1943|M.ulcerans_Agy99 GAVKEVSVS-AEPKVKVVIPVDPAGDGGLMDIVLSPTYSQDR--LMYAYI
MAV_3855|M.avium_104 GAVKEISVS-AEPKVKTVIPVDPSGDGGLMDIVLSPTYQQDR--LMYAYV
MLBr_01699|M.leprae_Br4923 GAVKEISVS-AEPKVKTVVSVDSSGDGGLMDIVLSPTYQQDR--LMYAYI
MAB_3333|M.abscessus_ATCC_1997 GVIKEIATS-AEPKIKTTIPVDPSGDGGLSDIVLSPSYQQDR--LMYAYI
MSMEG_1214|M.smegmatis_MC2_155 AAIKHLTAPGVVEQIGAVPGVAPRGEGGLLGLATAGAN-------VYAYL
TH_2267|M.thermoresistible__bu GMIKQLSEPGVVRDVGAVEGVAARGEGGLLGLATAGDT-------VFAYH
Mflv_5245|M.gilvum_PYR-GCK -AIRVVEDGVLRTDPLITIAVRTESERGIGGLAVDPDFEDNGRIYVAYIA
Mvan_1010|M.vanbaalenii_PYR-1 -AIRVVEDGVLREDPLITLAVRTESERGIEGLAVDPAFGTNGRIYVAYID
:. : . . * . .: *: .:. :
Mb3031|M.bovis_AF2122/97 STPTDNRVVRVADG----DIPKDILTGIPKGAAGNTG-ALIFTSPTTLVV
Rv3006|M.tuberculosis_H37Rv STPTDNRVVRVADG----DIPKDILTGIPKGAAGNTG-ALIFTSPTTLVV
MMAR_1706|M.marinum_M STPTDNRVIRIADG----DVPKDILTGIPKGPTGNTG-ALIFTSPTTLVV
MUL_1943|M.ulcerans_Agy99 STPTDNRVIRIADG----DVPKDILTGIPKGPTGNTG-ALIFTSPTTLVV
MAV_3855|M.avium_104 STPTDNRVIRVADG----DIPKDILTGIPKGATGNTG-ALMFTSPTTLVV
MLBr_01699|M.leprae_Br4923 STPTDNRVIRVADG----DIPKNILTGIPKGAVGNTG-ALIFTSPTTLVV
MAB_3333|M.abscessus_ATCC_1997 STPTDNRVVRVADG----DVPKDILTGIPKGPTGNAG-SLAFTSPTTLVV
MSMEG_1214|M.smegmatis_MC2_155 TTGGDNRVVQMRFADGALTNPTPILTGIPAG-SRHDGGRIAFGPDGKLYV
TH_2267|M.thermoresistible__bu TTATDNRVVRMDFDGQALGPPAPILTGIPTGGSRHDGGRIAFGPDGRLYV
Mflv_5245|M.gilvum_PYR-GCK AETTRNTLSRFVVDGNTASFDAQLVESTLTAAPNHHGGALAFGPDGKLYW
Mvan_1010|M.vanbaalenii_PYR-1 AETTRNTLSRFIVNGNTATFDTDLLVSTLAAAPNHHGGALGFGPDGALYW
: * : :. :: . . : * : * . *
Mb3031|M.bovis_AF2122/97 MTGDAGDPALAADPQSLAGKVLRIEQPTTIDQTPPTT-------ALSGIG
Rv3006|M.tuberculosis_H37Rv MTGDAGDPALAADPQSLAGKVLRIEQPTTIGQTPPTT-------ALSGIG
MMAR_1706|M.marinum_M MTGDAGSPEMAADPNSLAGKVLRIEQPTTIGQAPPTT-------ALAGVG
MUL_1943|M.ulcerans_Agy99 MTGDAGSPEMAADPNSLAGKVLRIEQPTTIGQAPPTT-------ALAGVG
MAV_3855|M.avium_104 LTGDAGNPALAADPKSLAGKVLRIEQPTTVGQAPPTT-------ALSGVG
MLBr_01699|M.leprae_Br4923 LTGDVDNPAMAADPKSLAGKLLRIEQPTTIGQAPPTT-------ALSSIG
MAB_3333|M.abscessus_ATCC_1997 QTGTGGNAGAANDPNSLAGKVIRIEQPTTVGQTPPTT-------AMSGLG
MSMEG_1214|M.smegmatis_MC2_155 ATGEIANPPLAQDRTSLAGKILRVNPDGSVPADNPFP---GSPVWSYGHR
TH_2267|M.thermoresistible__bu ATGEIGDPPLAQDRSSLAGKILRINPDGSIPADNPDP---QSPVWSFGHR
Mflv_5245|M.gilvum_PYR-GCK GVGDNASGSNAQNLANIHGKILRLNTDGTVPTDNPVLDGERTGIYAYGLR
Mvan_1010|M.vanbaalenii_PYR-1 GVGDNATGANAQNLGNIHGKILRLNTDGSVPVDNPVINGARTHIYAYGLR
.* * : .: **::*:: :: * .
Mb3031|M.bovis_AF2122/97 SGGGLCIDPVDGSLYVADRTPTADRLQRITKNSEVSTVWTWPDKPGVAG-
Rv3006|M.tuberculosis_H37Rv SGGGLCIDPVDGSLYVADRTPTADRLQRITKNSEVSTVWTWPDKPGVAG-
MMAR_1706|M.marinum_M SGGGMCIDPVDGSLYIADRTPTADRLQRITKNSEVSTVWTWPDKPGVAG-
MUL_1943|M.ulcerans_Agy99 SGGGMCIDPVDGSLYIADRTPTADRLQRITKNSEFSTVWTWPDKPGVAG-
MAV_3855|M.avium_104 SGGGLCTDPVDGSLYVADRTPTADRLQRITKTSDVSTVWTWPDKPGVAG-
MLBr_01699|M.leprae_Br4923 SGGGLCTDPVDDSLYVTDRTPTADRLQRITKNSKVSTVWTWPDKPGVAG-
MAB_3333|M.abscessus_ATCC_1997 DGGAMCVDAANGALYVTDRAPAGDRLQKLTKDGKISTVWTWPDKPGVAG-
MSMEG_1214|M.smegmatis_MC2_155 NIQGLAWDAAGRLWATEYGADRVDELNLIRPGANYG----WPEAEGRGG-
TH_2267|M.thermoresistible__bu NVQGLAWDPGGRLWATEYGADTWDELNLIEPGGNYG----WPVAEGRQE-
Mflv_5245|M.gilvum_PYR-GCK NPFRMTFTPTGELLVADVGAAAFEEVNNVVSGGNYG----WPSSEGVCTS
Mvan_1010|M.vanbaalenii_PYR-1 NPFRLTFTPTGQLLVADVGAASFEEVNLVTAGGNYG----WPSSEGLCTS
. : . . : :.:: : .. . ** *
Mb3031|M.bovis_AF2122/97 -CAAMDGTVLVNLINTKLTVAVRLAPSTGAVTGEPDVVR-KDTHAHAWAL
Rv3006|M.tuberculosis_H37Rv -CAAMDGTVLVNLINTKLTVAVRLAPSTGAVTGEPDVVR-KDTHAHAWAL
MMAR_1706|M.marinum_M -CAAMDGTVLVNLIETKLTVAVRLAASTGAVTGEPDVVR-KDTHAHAWAL
MUL_1943|M.ulcerans_Agy99 -CAAMDGTVLVNLIETKLTVAVRLAASTGAVTGEPDVVR-KDTHAHAWAL
MAV_3855|M.avium_104 -CAAMDGTVLVNLINTKMTVAVRLAPNTGAVTGEPDVVR-KDTHAHAWAL
MLBr_01699|M.leprae_Br4923 -CAAMDGTVLVNLISPKLTVAVRLAPSTGAVTGEPDVVR-KDTHVHAWAL
MAB_3333|M.abscessus_ATCC_1997 -CAALDSSVMVNLVFNQTTVVVRQNPETGAVTGDPEVLRQKEQHGHVRAL
MSMEG_1214|M.smegmatis_MC2_155 -IEGMVDPVVEWG-TGEASPSG-LAFYGGSLWVAALRGERLFQIPVAGDG
TH_2267|M.thermoresistible__bu -MPGMIDPVLQWR-PAEASPSG-LAFWDGALWIANLRGQRLLEVPVRADG
Mflv_5245|M.gilvum_PYR-GCK NCGGKIDPIYSYPRAGGAAITS-VAVRRGKVFIADTVKGWITVLTCTPGY
Mvan_1010|M.vanbaalenii_PYR-1 ACTGKTDPVFTYPRGSGAAITS-VAVYQGKVFIADLVQGWIKVLTCTPDY
. ..: : * :
Mb3031|M.bovis_AF2122/97 RMSPDGNVWGATVNKTAGDAEKLDDVVFPLFPQGGGFPRNNDDKT-----
Rv3006|M.tuberculosis_H37Rv RMSPDGNVWGATVNKTAGDAEKLDDVVFPLFPQGGGFPRNNDDKT-----
MMAR_1706|M.marinum_M RMSPDGNVWGATVNRTAGDAEKLDDVVFPLFPQGGGFPRNNDDKT-----
MUL_1943|M.ulcerans_Agy99 RMSPDGNVWGATVNRTAGDAEKLDDVVFPLFPQGGGFPRNNDDKT-----
MAV_3855|M.avium_104 RMSPDGNVWGATVNKTAGDAEKLDDVVFPLFPQGGGFPRNNDDKT-----
MLBr_01699|M.leprae_Br4923 RMSPDGNVWGATINKTAGDAEKLDDVVFPLFPRGGGFPRNNDDKT-----
MAB_3333|M.abscessus_ATCC_1997 KLSADGNVWGGSINKDFGTATDTEDVVFPLFPKGGGFPRANDDLD-----
MSMEG_1214|M.smegmatis_MC2_155 TVGTPVPLFVGQYGRLRTVVAAPDGSLWFTTSNRDGRGSVRAGDDRILQF
TH_2267|M.thermoresistible__bu ELGEPVSRFVGEYGRLRTVVAAPDGSLWFTTSNRDGRGVPRSGDDRILRY
Mflv_5245|M.gilvum_PYR-GCK TSCGEATTFDPDAGATVVLAEGPDGALYQLIYQPGSLVRIELADAAPEQV
Mvan_1010|M.vanbaalenii_PYR-1 TACGDVQTFDPAAGTTVVLAAGPDGNLYQLTYAPGELIAIKPAGGDADSE
: . . :. :: . .
Mb3031|M.bovis_AF2122/97 --
Rv3006|M.tuberculosis_H37Rv --
MMAR_1706|M.marinum_M --
MUL_1943|M.ulcerans_Agy99 --
MAV_3855|M.avium_104 --
MLBr_01699|M.leprae_Br4923 --
MAB_3333|M.abscessus_ATCC_1997 --
MSMEG_1214|M.smegmatis_MC2_155 RP
TH_2267|M.thermoresistible__bu RR
Mflv_5245|M.gilvum_PYR-GCK --
Mvan_1010|M.vanbaalenii_PYR-1 V-