For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNHAITHDIVITGGRVIDGTGRPAERADLAVTGDVITAIGMGPYQATTRIDATDRVVAPGFIDLHSHADF TVAGHPTAETAIAQGVTTLITGNCGQSTFPVRDQEEARLSTAFRDFPLSWEWTDAAGFGETVMAAGPAVN VGLQVGHNALRVAVMGQQDRAPDTRELREMCSLIEESAQHPGVVGFSTGLIYIPGLFAFTDELVALVSAA ASCDLLYSTHMRSETSELVEAVEEAIGTAERSGARLEISHLKAMGPENWGAVDKALELIDAARARGVDVA ADVYPYTASSTDLSSRVPRWAIDGGRPALLDRLADPVQRARIAEEMRARFGRDIDPEGVVIADLPPGEFD SYVGWSIARIGRDRGVDAAEVALDVLAAHAGSVTIVNHAMSEDDVRTVLAHPWVSVASDGWMLAPSGPGR PHPRSFGTFPRVLGRYVREQPTISLEEAIRKMTTLPASRIGITDRGSLTVGAAADIVVLDPHSIADTSTF DDPWQLARGVHTVLVNGTPTLLDGEFTKNHAGRLLQKRKPA
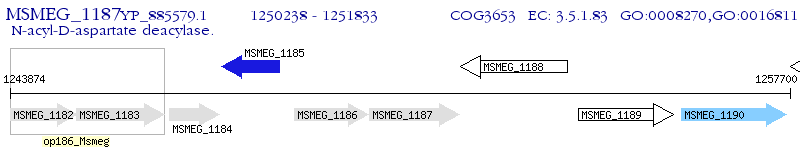
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1187 | - | - | 100% (531) | N-acyl-D-aspartate deacylase |
| M. smegmatis MC2 155 | MSMEG_3787 | - | 2e-82 | 36.29% (529) | D-aminoacylase |
| M. smegmatis MC2 155 | MSMEG_0883 | - | 2e-27 | 26.01% (596) | amidohydrolase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2937c | - | 2e-21 | 25.00% (544) | D-amino acid aminohydrolase |
| M. gilvum PYR-GCK | Mflv_0136 | - | 3e-37 | 27.62% (583) | amidohydrolase 3 |
| M. tuberculosis H37Rv | Rv2913c | - | 2e-21 | 25.00% (544) | D-amino acid aminohydrolase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2605c | - | 3e-25 | 23.95% (597) | hypothetical protein MAB_2605c |
| M. marinum M | MMAR_0213 | - | 2e-35 | 27.79% (601) | D-amino acid aminohydrolase |
| M. avium 104 | MAV_5277 | - | 3e-36 | 27.50% (600) | amidohydrolase family protein, putative |
| M. thermoresistible (build 8) | TH_3974 | - | 6e-37 | 27.35% (607) | PUTATIVE Amidohydrolase 3 |
| M. ulcerans Agy99 | MUL_2043 | - | 3e-24 | 23.88% (603) | D-amino acid aminohydrolase |
| M. vanbaalenii PYR-1 | Mvan_3037 | - | 1e-34 | 27.47% (597) | amidohydrolase 3 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0213|M.marinum_M --------------MFDLKIVGGTVVDGTGAARYRADIGIKDGKIVDVVR
MAV_5277|M.avium_104 --------------MFDLKITGGTVVDGTGAQRYRADVGIRDGKIVDVVR
TH_3974|M.thermoresistible__bu -------------MVFDLKITGGTVVDGTGADRFLGDIGIAGGTIVEVRR
Mvan_3037|M.vanbaalenii_PYR-1 --------------MLDLKITGGTVVDGTGADRFRADIGVKDGRIVEVRR
Mflv_0136|M.gilvum_PYR-GCK -------------MTYDLVIRGGTVVDGLGGEPFTADVAVTDGIIREVGA
Mb2937c|M.bovis_AF2122/97 MLAWRQLNDLEETVTYDVIIRDGLWFDGTGNAPLTRTLGIRDGVVATVAA
Rv2913c|M.tuberculosis_H37Rv MLAWRQLNDLEETVTYDVIIRDGLWFDGTGNAPLTRTLGIRDGVVATVAA
MUL_2043|M.ulcerans_Agy99 -------------MTYDVIIRDGLWFDGTGSAPQTRTLGIRDGMVATVSA
MAB_2605c|M.abscessus_ATCC_199 MVPCS-------DAQFDTVIRNGRWFDGTGAPSAIRNIGIKDGHVAAVSA
MSMEG_1187|M.smegmatis_MC2_155 ---------MNHAITHDIVITGGRVIDGTGRPAERADLAVTGDVITAIGM
* * .* .** * :.: .. : :
MMAR_0213|M.marinum_M R-GTDDPGLG-GESAETIDATGQVVAPGFVDIHTHYDGQVSWDSTLEPSS
MAV_5277|M.avium_104 ADGSEAGGLATAEAAETIDATGKIVAPGFVDIHTHYDGQVSWDSLLEPSS
TH_3974|M.thermoresistible__bu R-GPGDPPLE-GAAAETLDATGTIVAPGFVDIHTHYDGQVSWDDLLEPSS
Mvan_3037|M.vanbaalenii_PYR-1 R-DGDDPGLQ-TEAAHQIDATGRIVAPGFVDVHTHYDGQVSWDETLEPSS
Mflv_0136|M.gilvum_PYR-GCK V---------GSAGAREIDATGLLVTPGFVDLHTHYDGQAIWSDRLTPSS
Mb2937c|M.bovis_AF2122/97 G------ALDETGCPEVVDAAGKWVVPGFIDVHTHYDAEVLLDPGLRESV
Rv2913c|M.tuberculosis_H37Rv G------ALDETGCPEVVDAAGKWVVPGFIDVHTHYDAEVLLDPGLRESV
MUL_2043|M.ulcerans_Agy99 G------PLDETGCAQVIDAAGKWVMPGFIDVHTHYDAEVLLDPGLRESV
MAB_2605c|M.abscessus_ATCC_199 D------DLDETHCPNIIDASGKWVLPGLVDIHTHYDIEVLAAPALSESV
MSMEG_1187|M.smegmatis_MC2_155 G---------PYQATTRIDATDRVVAPGFIDLHSHADFTVAGHPTAETAI
. :**:. * **::*:*:* * . :
MMAR_0213|M.marinum_M GHGVTTVVTGNCGVGFAPVRPGSEDWLIRLMEGVEDIPGTALTEGIT--W
MAV_5277|M.avium_104 GHGVTTVVTGNCGVGFAPVRPGTEQWLIELMEGVEDIPGTALTEGIT--W
TH_3974|M.thermoresistible__bu GHGVTTVVMGNCGVGFAPVRPGREEWLIELMEGVEDIPGAALSEGIS--W
Mvan_3037|M.vanbaalenii_PYR-1 LHGVTTVVSGNCGVGFAPVRPGREQWLIELMEGVEDIPGSALAEGIT--W
Mflv_0136|M.gilvum_PYR-GCK AHGVTTAVMGNCGVGFAPCRPADHDVLVDVMAGVEDIPGVVMVDGLP--W
Mb2937c|M.bovis_AF2122/97 RHGVTTVLLGNCSLSTVYANSEDAADLFSRVEAVPREFVLGALRDNQ---
Rv2913c|M.tuberculosis_H37Rv RHGVTTVLLGNCSLSTVYANSEDAADLFSRVEAVPREFVLGALRDNQ---
MUL_2043|M.ulcerans_Agy99 RHGVTTVLLGNCSLSTVYAGAEDAADLFSRVEAVPRQFVLGALSAAKSER
MAB_2605c|M.abscessus_ATCC_199 RHGVTTVLLGSCSLSVVHVDGVDAGDIFGRVEAIPRDHVIQVVDRYK---
MSMEG_1187|M.smegmatis_MC2_155 AQGVTTLITGNCGQSTFPVRDQEEARLSTAFRDFP----------LS--W
:**** : *.*. . : . .
MMAR_0213|M.marinum_M GWESYPQYLDTIGKQKFSIDVGSQIAHGSVRAYTMGERGARNE--PATAE
MAV_5277|M.avium_104 GWETYPEYLDAIGKQKFAIDVGSQVAHGAIRAYAMGERGARNE--PATPD
TH_3974|M.thermoresistible__bu GWESYADYLDVIGGRKLAIDVGSQIAHGTVRAYAMGERGARNE--PATPD
Mvan_3037|M.vanbaalenii_PYR-1 QWESFPEYLDAIEKRSLAIDYGTQIAHGAVRGYAMGERGANNE--EATAD
Mflv_0136|M.gilvum_PYR-GCK QWETFPEFLDAVASRQRDIDVAAFLPHSPLRVYVMGRRGVDRE--PATDE
Mb2937c|M.bovis_AF2122/97 TWSTPAEYIEAIDALPLGPNVSSLLGHSDLRTAVLGLDRATDDTVRPTEA
Rv2913c|M.tuberculosis_H37Rv TWSTPAEYIEAIDALPLGPNVSSLLGHSDLRTAVLGLDRATDDTVRPTEA
MUL_2043|M.ulcerans_Agy99 AWSNAAEYITAIDSLPLGPNVSSLLGHSDLRATVLGLERATDASVRPSEA
MAB_2605c|M.abscessus_ATCC_199 TWTNAGQYVAALESLPLGPNVAAFLGHSDIRAAAMGLDRATRKDERPTAA
MSMEG_1187|M.smegmatis_MC2_155 EWTDAAGFGETVMAAGPAVNVGLQVGHNALRVAVMGQQDRAPD----TRE
* : .: : . : *. :* .:* :
MMAR_0213|M.marinum_M DIAAMGRLVTEAIEAGALGFSTSRTLAHRAMD----GEPVPGTFAAEDEL
MAV_5277|M.avium_104 DIEAMGRLVREAIEAGALGFSTSRTMGHRAMD----GEPVPGTFAAEEEL
TH_3974|M.thermoresistible__bu DIAAMSRIVREAAEAGALGFSSSRTLAHRAID----GEPVPGTFAAEDEL
Mvan_3037|M.vanbaalenii_PYR-1 DIAAMARIVREAIEAGALGFSTSRTEAHRAID----GQAVPGTYAAEAEL
Mflv_0136|M.gilvum_PYR-GCK DLEMMRKLAAEAVHAGALGFASSRFTLHKTES----GQPIPSYNADYAEI
Mb2937c|M.bovis_AF2122/97 ELAKMAKLLDEALEAGMLGMSGMDAAIDKLDGDRFRSRALPSTFATWRER
Rv2913c|M.tuberculosis_H37Rv ELAKMAKLLDEALEAGMLGMSGMDAAIDKLDGDRFRSRALPSTFATWRER
MUL_2043|M.ulcerans_Agy99 ELGRMATLLDEALDAGMLGMSGMDSAIDKLDGERFRSRALPSTFATWRER
MAB_2605c|M.abscessus_ATCC_199 EQAQMDRWLTEALDAGFVGMSSQQLLFDKLDGDVCRSRTLPSTYAKPREL
MSMEG_1187|M.smegmatis_MC2_155 LREMCSLIEESAQHPGVVGFSTG-------------LIYIPGLFAFTDEL
.* ..* :*:: :*. * *
MMAR_0213|M.marinum_M FGLGRAMAAGGQAVFELAPQGAAGEDIVGP-RKELEWMRRLAGEIDRPVS
MAV_5277|M.avium_104 FGLGRAMAAGGQAVFELAPQGAAGEDIIGP-KKELDWMRRLSGEIDRPVS
TH_3974|M.thermoresistible__bu FALGRALADGGGAVFELAPQGAAGEDIVAP-GKELEWMRRLGAEIDCALS
Mvan_3037|M.vanbaalenii_PYR-1 FGLGRAMAAGGQAVFEVAPQGTAGESPADACMREMEWMAKLSDDISRPVS
Mflv_0136|M.gilvum_PYR-GCK EAIARGVQDGGGGLIQFVPDLVAGD--YEP---VLQTVFDAAADVGLPVT
Mb2937c|M.bovis_AF2122/97 RKLISVLRHRGR-ILQSAPD---VDNPVSALLFFLASSRIFNRRKGVRMS
Rv2913c|M.tuberculosis_H37Rv RKLISVLRHRGR-ILQSAPD---VDNPVSALLFFLASSRIFNRRKGVRMS
MUL_2043|M.ulcerans_Agy99 RRLIKVLRKRGR-ILQSAPN---VDKPASALLFFLASSRILNRRKGVRMS
MAB_2605c|M.abscessus_ATCC_199 RRLKAQLRQRNR-VLQSGPD---IQNPLNVLSQAVQSLGILRRP--LSTS
MSMEG_1187|M.smegmatis_MC2_155 VALVSAAASCDLLYSTHMRS--ETSELVEAVEEAIGTAERSGAR--LEIS
: . . . : :
MMAR_0213|M.marinum_M FALIQVDADPNLWREQLELSASAHAAGSRLHPQIAARPFGMMIGFQG-HH
MAV_5277|M.avium_104 FALIQVDADPNLWREMLDLSADAHAAGARLYPQIAARPFGMMIGFQG-HH
TH_3974|M.thermoresistible__bu FALIQVDADPNLWREQLEISAAAHEAGSRLYPQVAARPFGMLLGFPG-HH
Mvan_3037|M.vanbaalenii_PYR-1 FTLIQASHAPDLWRQQLDRAEKAHENGIELYAQFAARPFGMLLGFPG-YH
Mflv_0136|M.gilvum_PYR-GCK FTLAVGNAGDPFFLDALTMVEKANANGGDITAQIFPRPIGLVLGLELSGN
Mb2937c|M.bovis_AF2122/97 MLVSADAKSMPLAVHVFGLGTRVLNKLLGSQVRFQHLPVPFELYSDG---
Rv2913c|M.tuberculosis_H37Rv MLVSADAKSMPLAVHVFGLGTRVLNKLLGSQVRFQHLPVPFELYSDG---
MUL_2043|M.ulcerans_Agy99 LLVSADSKSMPMAVRTFGLGARLLNKLLAASVRFQHLPVPFELYSDG---
MAB_2605c|M.abscessus_ATCC_199 LLSAADIKANPYAIRAMGPVASLVNKLGG-NFRWQHLPVPFEVYADG---
MSMEG_1187|M.smegmatis_MC2_155 HLKAMGPENWGAVDKALELIDAARARGVDVAADVYPYTASSTDLSSR---
: .
MMAR_0213|M.marinum_M AFTHRPTYRRLKADCTRDELAQRLADPAVKAAILSEDDLPIDPNVLFDG-
MAV_5277|M.avium_104 GFSHRPTYRRLAAECSREELAQRLADPAVKAAILGEDDLPVDPTLLFDG-
TH_3974|M.thermoresistible__bu AFTHRPTYRRLAAECDREELAARLAEPAVRAAILAEDDLPPDPDVLFDG-
Mvan_3037|M.vanbaalenii_PYR-1 AFTHRPTFRAVSAGSTRDELAARLADPAVKAAILAEEDLPPTPGALLDG-
Mflv_0136|M.gilvum_PYR-GCK PFVMYPSYREIAALP-LAERVAEMRKPEVRERILN--DTPAS-----DGH
Mb2937c|M.bovis_AF2122/97 --IDLPVFEEFGAGTAALHLRDQLQ----RNELLADRSYRRSFRREFDRI
Rv2913c|M.tuberculosis_H37Rv --IDLPVFEEFGAGTAALHLRDQLQ----RNELLADRSYRRSFRREFDRI
MUL_2043|M.ulcerans_Agy99 --IDLPVFEEFGAGTAALHLREQVQ----RNQLLADESYRSEFRREFDRT
MAB_2605c|M.abscessus_ATCC_199 --IDLVIFEEFGAGAAALHLRDEVE----RNELLRDEKYRRRFRKEYES-
MSMEG_1187|M.smegmatis_MC2_155 -------VPRWAIDGGRPALLDRLADPVQRARIAEEMRARFGRDIDPEG-
.: : : :
MMAR_0213|M.marinum_M MSAFVQYSLGRVYALGDPPDYEPTRDRTVAAIAQARG-QDPLATLYDLML
MAV_5277|M.avium_104 MFALVQHSLHRLYALGDPPDYEPTPDRTVAAIAEARG-EDPLATLYDLML
TH_3974|M.thermoresistible__bu MFAFAQHAVDRLYPLGDPPDYEPTPDRTVTAIAAERG-VDPLTAMYDLML
Mvan_3037|M.vanbaalenii_PYR-1 LFALAQYSTDRIYAIGDPPDYEPTPDKTVAAIAAARG-QKPLETMYDLML
Mflv_0136|M.gilvum_PYR-GCK PLMFAAQAWNYMFPLTDPPNYEPSPSDSIGARARARG-VSPLEEAYDRLL
Mb2937c|M.bovis_AF2122/97 KLGPSLWHRDFHDAVIVECPDKSLIGKSFGAIADERG-LHPLDAFLDVLV
Rv2913c|M.tuberculosis_H37Rv KLGPSLWHRDFHDAVIVECPDKSLIGKSFGAIADERG-LHPLDAFLDVLV
MUL_2043|M.ulcerans_Agy99 KLGPSLWHKDFHDAVIVECPDESLIGKSFGAIADERG-LHPLDAFLDVLV
MAB_2605c|M.abscessus_ATCC_199 KFGVRVWHRDFFDAEIVSCPDKSVLGKSFGQVGMDRGGLHPVDAFLDLVL
MSMEG_1187|M.smegmatis_MC2_155 ----------VVIADLPPGEFDSYVGWSIARIGRDRG-VDAAEVALDVLA
. .. . :. . ** . * :
MMAR_0213|M.marinum_M ESNASAMLMLPLFNYADGNCDAIREMMLHPAGVLGLSDGGAHCGMICDAS
MAV_5277|M.avium_104 DADATNMLMLPLFNYADGNCDAIREMLLHPAGVLGLSDGGAHCGMICDAS
TH_3974|M.thermoresistible__bu ENGAGAMLLLPLYNYADGNHDAIREMLTHPAGVLGLSDGGAHCSMICDAS
Mvan_3037|M.vanbaalenii_PYR-1 EADAGAMLMLPFFNYAEGDHRAIYEMMQSPAAVSGLSDGGAHCGLICDAS
Mflv_0136|M.gilvum_PYR-GCK DDDGHAMLLVTLANFRDGTLDTVAELIRRDDVVLGLGDGGAHYGMICDAS
Mb2937c|M.bovis_AF2122/97 DNGERNVRWTTIVANHR--PNQLNKLAAEPSVHMGFSDAGAHLRNMAFYN
Rv2913c|M.tuberculosis_H37Rv DNGERNVRWTTIVANHR--PNQLNKLAAEPSVHMGFSDAGAHLRNMAFYN
MUL_2043|M.ulcerans_Agy99 ENGERNVRWTTVVANHR--PKQLNKLAAEPGVHMGFSDAGAHLRNMAFYN
MAB_2605c|M.abscessus_ATCC_199 EHG-TAMRWRTTISNHR--PEVLKQLAQDPGVQIGFSDAGAHLRNMAFYN
MSMEG_1187|M.smegmatis_MC2_155 AHAGSVTIVNHAMSEDDVRTVLAHPWVSVASDGWMLAPSGPGRPHPRSFG
:. .*. .
MMAR_0213|M.marinum_M YPTFLLT-HWARDRKRGEKLPLEYVIRKQARDTAHLFGLTDRGTIEPGKK
MAV_5277|M.avium_104 YPTFLLT-HWARDRSRGDKLALEYVIRKQSRDTAHLFGLTDRGTIEPGKK
TH_3974|M.thermoresistible__bu YPTFLLT-HWARDRSRGDRLPLEYVIRKQAHDTAQLFGLTDRGTIAVGKK
Mvan_3037|M.vanbaalenii_PYR-1 YPTFLLT-HWARDRHRGPKFSLEYVVRKQTLDTATLFGLSDRGVIAAGKK
Mflv_0136|M.gilvum_PYR-GCK FPTYMLT-HWVRDRPSG-RLTVPEAVRELTSVPARVAGLGDRGRIAVGYK
Mb2937c|M.bovis_AF2122/97 FGLRLLKRARDADRAGQPFLSIERAVYRLTGELAEWFGIG-AGTLRQGDR
Rv2913c|M.tuberculosis_H37Rv FGLRLLKRARDADRAGQPFLSIERAVYRLTGELAEWFGIG-AGTLRQGDR
MUL_2043|M.ulcerans_Agy99 FPLRLLKRTWDAERAGKPFLSLQQAVYRLTGELAEWFGVD-AGTLREGDR
MAB_2605c|M.abscessus_ATCC_199 FGLRLLRHVREAQQAGRPFMTVEQAVHRLTGELADWYQID-AGHLRIGDR
MSMEG_1187|M.smegmatis_MC2_155 TFPRVLG----RYVREQPTISLEEAIRKMTTLPASRIGITDRGSLTVGAA
:* :.: .: . : * : * : *
MMAR_0213|M.marinum_M ADLNVIDLEALRLHPAAMAYDLPAGGNRILQGASGYAATIVSGAVTR---
MAV_5277|M.avium_104 ADINVIDMDALRLHPAAMAFDLPAGGNRILQGASGYAATIVSGTVTR---
TH_3974|M.thermoresistible__bu ADLNVIDLDALTLHPPRMVYDLPAGGHRLVQGASGYVATIVGGVVTR---
Mvan_3037|M.vanbaalenii_PYR-1 ADINVIDMDALRLDVPRMVYDLPADGRRLLQGASGYDATVVNGVVTR---
Mflv_0136|M.gilvum_PYR-GCK ADLNVIDADTLRLHRPVIVKDLPGGGRRLDQTADGYVATIVSGEVIA---
Mb2937c|M.bovis_AF2122/97 ADFAVIDPTHLDESVDGYHEEAVPYYGGLRRMVNRNDATVVATGVGGTVV
Rv2913c|M.tuberculosis_H37Rv ADFAVIDPTHLDESVDGYHEEAVPYYGGLRRMVNRNDATVVATGVGGTVV
MUL_2043|M.ulcerans_Agy99 ADFVVIDPAHLDESVDGYHEAQVPFYGGLSRMVNRNDATVIATGVAGAVV
MAB_2605c|M.abscessus_ATCC_199 ADLFVIDPEKLDATLTAYAEESVDAYGGLSRMVNRNDATVSTVLIGGQPV
MSMEG_1187|M.smegmatis_MC2_155 ADIVVLDPHSIADTST--FDDPWQLARGVHTVLVNGTPTLLDGEFTKN--
**: *:* : : .*: .
MMAR_0213|M.marinum_M ----------RGGVDTGARPGRLVRGAR--------------
MAV_5277|M.avium_104 ----------RNDVDTGARPGRLVRGAR--------------
TH_3974|M.thermoresistible__bu ----------RDGVDTGARPGRLVRGAR--------------
Mvan_3037|M.vanbaalenii_PYR-1 ----------RHGVDTGARPGRLLRGVR--------------
Mflv_0136|M.gilvum_PYR-GCK ----------ENGEPTDARPGALIRGRRPAPVA---------
Mb2937c|M.bovis_AF2122/97 FRGGQFGGQFRDGYGQNVKSGRYLRAGELGAALSRSA-----
Rv2913c|M.tuberculosis_H37Rv FRGGQFGGQFRDGYGQNVKSGRYLRAGELGAALSRSA-----
MUL_2043|M.ulcerans_Agy99 FG----TGQFRDGYGQTVRSGRYLRAGQRYTAASVSA-----
MAB_2605c|M.abscessus_ATCC_199 VV----DGQLRDLLGK-RRTGRFLPAGAKARAVPVDTAPAPH
MSMEG_1187|M.smegmatis_MC2_155 ------------------HAGRLLQKRKPA------------
:.* :