For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGPTVLFLCTHNAGRSQMALGYFTHLAGDGAVAWSGGSEPAAEINPVAVAAMAEVGIDITGEYPKPWTDE IVRVADVVITMGCGDTCPVYPGKRYENWELPDPAGQALDAVRPIRDDIEHRVRRLLADLGVATIR*
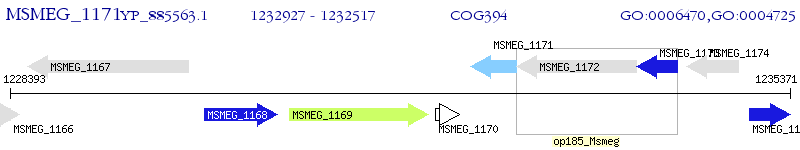
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1171 | - | - | 100% (136) | low molecular weight phosphotyrosine protein phosphatase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2676 | arsC | 2e-43 | 63.78% (127) | arsenic-transport integral membrane protein ArsC |
| M. gilvum PYR-GCK | Mflv_1659 | - | 6e-59 | 78.29% (129) | protein tyrosine phosphatase |
| M. tuberculosis H37Rv | Rv2643 | arsC | 3e-43 | 63.78% (127) | arsenic-transport integral membrane protein ArsC |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2568 | - | 8e-56 | 74.24% (132) | putative arsenate reductase or tyrosine phosphatase |
| M. marinum M | MMAR_2055 | arsC | 2e-43 | 63.85% (130) | arsenic-transport integral membrane protein ArsC |
| M. avium 104 | MAV_1496 | - | 6e-61 | 77.61% (134) | low molecular weight phosphotyrosine protein phosphatase |
| M. thermoresistible (build 8) | TH_1049 | arsC | 1e-40 | 60.63% (127) | PROBABLE ARSENIC-TRANSPORT INTEGRAL MEMBRANE PROTEIN ARSC |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3221 | - | 2e-63 | 84.50% (129) | protein tyrosine phosphatase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1659|M.gilvum_PYR-GCK --------------------------------------------------
MAV_1496|M.avium_104 --------------------------------------------------
Mvan_3221|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1171|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2568|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2676|M.bovis_AF2122/97 MTETVTRTAAPAVVGKLSTLDRFLPVWIGSAMAAGLLLGRWIPGLHTALE
Rv2643|M.tuberculosis_H37Rv MTETVTRTAAPAVVGKLSTLDRFLPVWIGSAMAAGLLLGRWIPGLHTALE
MMAR_2055|M.marinum_M MTGTVDHQPAPAVVGKLSTLDRFLPVWIGLAMAAGLLLGRWIPGLNTALE
TH_1049|M.thermoresistible__bu --------------------------------------------------
Mflv_1659|M.gilvum_PYR-GCK --------------------------------------------------
MAV_1496|M.avium_104 --------------------------------------------------
Mvan_3221|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1171|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2568|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2676|M.bovis_AF2122/97 GVQLDGISLPIALGLLIMMYPVLAKVRYDRLDTVTGDRKLLLSSLLLNWV
Rv2643|M.tuberculosis_H37Rv GVQLDGISLPIALGLLIMMYPVLAKVRYDRLDTVTGDRKLLLSSLLLNWV
MMAR_2055|M.marinum_M KVQVDGISLPIALGLLIMMYPVLAKVRYDRLDSVTGDRKLLISSLLLNWV
TH_1049|M.thermoresistible__bu --------------------------------------------------
Mflv_1659|M.gilvum_PYR-GCK --------------------------------------------------
MAV_1496|M.avium_104 --------------------------------------------------
Mvan_3221|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1171|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2568|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2676|M.bovis_AF2122/97 LGPALMFALAWLLLADLPEYRTGLIIVGLARCIAMVIIWNDLACGDREAA
Rv2643|M.tuberculosis_H37Rv LGPALMFALAWLLLADLPEYRTGLIIVGLARCIAMVIIWNDLACGDREAA
MMAR_2055|M.marinum_M AGPALMFALAWLLLPDLPEYRTGLIIVGLARCIAMVIIWNDLARGDREAA
TH_1049|M.thermoresistible__bu --------------------------------------------------
Mflv_1659|M.gilvum_PYR-GCK --------------------------------------------------
MAV_1496|M.avium_104 --------------------------------------------------
Mvan_3221|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1171|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2568|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2676|M.bovis_AF2122/97 AVLVALNSIFQVAMFAALGWFYLSVLPGWLGLEQTTIATSPWQIAKSVLI
Rv2643|M.tuberculosis_H37Rv AVLVALNSIFQVAMFAALGWFYLSVLPGWLGLEQTTIATSPWQIAKSVLI
MMAR_2055|M.marinum_M AVLVALNSIFQVAMFAVLGWFYLSVLPGWLHLTQTTISTSPWQIAKSVLI
TH_1049|M.thermoresistible__bu --------------------------------------------------
Mflv_1659|M.gilvum_PYR-GCK --------------------------------------------------
MAV_1496|M.avium_104 --------------------------------------------------
Mvan_3221|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1171|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2568|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2676|M.bovis_AF2122/97 FLGIPLLAGYLSRRIGEKTKGRNWYESRFLPKVGPWALYGLLFTIVILFA
Rv2643|M.tuberculosis_H37Rv FLGIPLLAGYLSRRIGEKTKGRNWYESRFLPKVGPWALYGLLFTIVILFA
MMAR_2055|M.marinum_M FLGIPLLAGYLSRRLGERAKGRGWYESRFLPKVGPWALYGLLFTIVILFA
TH_1049|M.thermoresistible__bu --------------------------------------------------
Mflv_1659|M.gilvum_PYR-GCK -----------------------MTDSIAPDTHPHLRHDLSIDQRLALKA
MAV_1496|M.avium_104 -----------------------MTDSPAS-LAAHVRRDISIDQQLALRT
Mvan_3221|M.vanbaalenii_PYR-1 -----------------------MTDTPVT---HQLRRDLPIDQQLALKA
MSMEG_1171|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2568|M.abscessus_ATCC_1997 -----------------------MTD------------------------
Mb2676|M.bovis_AF2122/97 LQGDQITGRPLDVARIALPLLAYFAIMWVGGYLLGAALRLGYRRTTTLAF
Rv2643|M.tuberculosis_H37Rv LQGDQITGRPLDVARIALPLLAYFAIMWVGGYLLGAALRLGYRRTTTLAF
MMAR_2055|M.marinum_M LQGDQITNRPQDVARIALPLLVYFAVMWGGGYLLGAALHLGYQRTTTMAF
TH_1049|M.thermoresistible__bu --------------------------------------------------
Mflv_1659|M.gilvum_PYR-GCK AATHLETEFDGMYGMETIERFLHSSYDQFAGRASIPNFLPLLAERFARQR
MAV_1496|M.avium_104 ASTRLHDDFAEHFGVETIERFLHSSYDQFAGRATVPNFLPLLAERWARQR
Mvan_3221|M.vanbaalenii_PYR-1 AANQLQTEFGDTIGAETIERFLHSSYAHFAGRAKVPNFLPLLAERYARQQ
MSMEG_1171|M.smegmatis_MC2_155 ----------------------------MTG-------------------
MAB_2568|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2676|M.bovis_AF2122/97 TAASNNFELAIAVAIATYGATSGQALAGVVGPLIEVPVLVGLVYVSLALR
Rv2643|M.tuberculosis_H37Rv TAASNNFELAIAVAIATYGATSGQALAGVVGPLIEVPVLVGLVYVSLALR
MMAR_2055|M.marinum_M TAAGNNFELAIAVAIATYGATSGQALAGVVGPLIEVPVLVGLVYVSLALR
TH_1049|M.thermoresistible__bu --------------MTSDSAT-----------------------------
Mflv_1659|M.gilvum_PYR-GCK LRALAKIEGKHHDGKPTVLFLCTHNAGRSQMAMGFFTHLAGDAAVAWSGG
MAV_1496|M.avium_104 LTALARVEGKVTDTKPTVLFLCTHNAGRSQMALGYFNHLAGDQGVAWSGG
Mvan_3221|M.vanbaalenii_PYR-1 LSALARVEAKIPDGKPTVLFLCTHNAGRSQMALGFLTHLAGDKVVGWSGG
MSMEG_1171|M.smegmatis_MC2_155 ---------------PTVLFLCTHNAGRSQMALGYFTHLAGDGAVAWSGG
MAB_2568|M.abscessus_ATCC_1997 --------------KPTVLFLCTHNAGRSQMALGYFTNLARGNAVGMSGG
Mb2676|M.bovis_AF2122/97 NRLAGPN-ATHDADKPSVLFVCVHNAGRSQMAAGLLTHLAGDRIEVRSAG
Rv2643|M.tuberculosis_H37Rv NRLAGPN-ATHDADKPSVLFVCVHNAGRSQMAAGLLTHLAGDRIEVRSAG
MMAR_2055|M.marinum_M NRYYGPHRRKHDMIKPSVLFVCVHNAGRSQMAAGWLRHLAGDRIEVRSAG
TH_1049|M.thermoresistible__bu -----------DNPTPSVLFVCVHNAGRSQMAAGFLTALAGGRIEVRSAG
*:***:*.********* * : ** . *.*
Mflv_1659|M.gilvum_PYR-GCK SEPGNEVNPAAVAAMAECNIDISREYPKPWTDEIVRAADVIITMGCGDAC
MAV_1496|M.avium_104 SEPGNEINPAAVQAMAEVGIDITREFPKPWTDEIVQASDVVVTMGCGDAC
Mvan_3221|M.vanbaalenii_PYR-1 SEPGHEINPAAVAAMAEVGIDITGEFPKPWTEEIVRAADVVVTMGCGDAC
MSMEG_1171|M.smegmatis_MC2_155 SEPAAEINPVAVAAMAEVGIDITGEYPKPWTDEIVRVADVVITMGCGDTC
MAB_2568|M.abscessus_ATCC_1997 SEPADQINPAAIAVMHEVGIDITTEYPKPWSVETVQAADVVVTMGCDDSC
Mb2676|M.bovis_AF2122/97 TEPAGQVNPTAVAAMAEMGIDITANAPTLLTGGQVQSSDVVITMGCGDAC
Rv2643|M.tuberculosis_H37Rv TEPAGQVNPTAVAAMAEMGIDITANAPTLLTGGQVQSSDVVITMGCGDAC
MMAR_2055|M.marinum_M TEPADRINPVAVAAMAELGIDITANTPEVLTGAWVQTSDVVITMGCGDAC
TH_1049|M.thermoresistible__bu SAPADQLNPAAVEAMAEVGIDISTHQPKVLTTDAVQASDVVITMGCGDTC
: *. .:**.*: .* * .***: . * : *: :**::****.*:*
Mflv_1659|M.gilvum_PYR-GCK PVFPGLRYEEWKLEDPHGQGVDAVRPIRDEIERRVRALLSELD----VPV
MAV_1496|M.avium_104 PFFPGKRYENWDLTDPAGQGVDVVRPIRDEIESRVRRLLAELH----ITP
Mvan_3221|M.vanbaalenii_PYR-1 PVFPGKRYENWELTDPAGQGLEAVRPIRDDVEERVRRLLADLQ----VPV
MSMEG_1171|M.smegmatis_MC2_155 PVYPGKRYENWELPDPAGQALDAVRPIRDDIEHRVRRLLADLG----VAT
MAB_2568|M.abscessus_ATCC_1997 PYFPGKRYENWELADPAGQSVEAIRPIRDDIEQRVRELLISLG----VAG
Mb2676|M.bovis_AF2122/97 PYFPGASYRNWKLPDPAGQPLDVVRMIRDDIADRVQALIAEL-----LAT
Rv2643|M.tuberculosis_H37Rv PYFPGVSYRNWKLPDPAGQPLDVVRMIRDDIADRVQALIAEL-----LAT
MMAR_2055|M.marinum_M PYFPGVSYRDWTLADPAGQPLDVVRPIRDDIGERVRALIAEL-----LNE
TH_1049|M.thermoresistible__bu PIFPGRSYRDWVLDDPAGKDLDDVRRIRDDIKARVEDLIAELDPPPDLNA
* :** *.:* * ** *: :: :* ***:: **. *: .* :
Mflv_1659|M.gilvum_PYR-GCK QS-------------
MAV_1496|M.avium_104 VS-------------
Mvan_3221|M.vanbaalenii_PYR-1 AVR------------
MSMEG_1171|M.smegmatis_MC2_155 IR-------------
MAB_2568|M.abscessus_ATCC_1997 VVEP-----------
Mb2676|M.bovis_AF2122/97 AKTR-----------
Rv2643|M.tuberculosis_H37Rv AKTR-----------
MMAR_2055|M.marinum_M HPDTAQICRG-----
TH_1049|M.thermoresistible__bu RDRPVPATRAGTTTR