For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MANRPLIGLTTYRESIRWGKWHMPALFTPETYVSAVDRAGGEPLLLPTALGHARSAVLALVDGLLVVGGA DIAAENYGERTQPETDPRPVRDRGELEALDYALGNDLPVLGVCRGMQLINVHAGGSLIQHLPTQVGHHEH LRTPGEFGSHPVRVDQASRISTVVGPGPVVTTYHHQGIGRLGSGLKAVAWADDGVIEAVESTTHSYVIGV QWHAERDDGTPLIEAFVEQCARRKAKWP
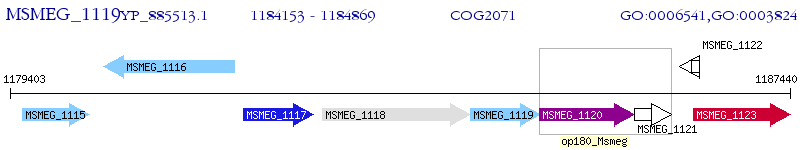
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1119 | - | - | 100% (238) | glutamine amidotransferase, class I |
| M. smegmatis MC2 155 | MSMEG_2596 | - | 6e-46 | 42.79% (229) | peptidase C26 |
| M. smegmatis MC2 155 | MSMEG_6691 | - | 1e-30 | 38.52% (244) | glutamine amidotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2884c | - | 3e-43 | 41.13% (231) | amidotransferase |
| M. gilvum PYR-GCK | Mflv_4064 | - | 4e-39 | 40.89% (225) | peptidase C26 |
| M. tuberculosis H37Rv | Rv2859c | - | 3e-43 | 41.13% (231) | amidotransferase |
| M. leprae Br4923 | MLBr_01573 | - | 8e-47 | 43.78% (217) | putative amidotransferase |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1844 | - | 3e-46 | 40.43% (235) | amidotransferase |
| M. avium 104 | MAV_3717 | - | 1e-42 | 43.46% (214) | class I glutamine amidotransferase, putative |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2093 | - | 2e-44 | 39.57% (235) | amidotransferase |
| M. vanbaalenii PYR-1 | Mvan_2279 | - | 1e-40 | 41.26% (223) | peptidase C26 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2884c|M.bovis_AF2122/97 MDLSASRSDGGDPLRPASPRLRSPVSDGGDPLRPASPRLRSPVSDGGDPL
Rv2859c|M.tuberculosis_H37Rv MDLSASRSDGGDPLRPASPRLRSPVSDGGDPLRPASPRLRSPVSDGGDPL
MAV_3717|M.avium_104 --------------------------------------------------
MLBr_01573|M.leprae_Br4923 --------------------------------------------------
MMAR_1844|M.marinum_M --------------------------------------------------
MUL_2093|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4064|M.gilvum_PYR-GCK ----------------------------------MSVS------------
Mvan_2279|M.vanbaalenii_PYR-1 ----------------------------------MNGSDPGASSRKLAQN
MSMEG_1119|M.smegmatis_MC2_155 --------------------------------------------------
Mb2884c|M.bovis_AF2122/97 RPASPRLRSPLGASRPVVGLTAYLEQVRTGVWDIPAGYLPADYFEGITMA
Rv2859c|M.tuberculosis_H37Rv RPASPRLRSPLGASRPVVGLTAYLEQVRTGVWDIPAGYLPADYFEGITMA
MAV_3717|M.avium_104 -----------------MGLTAYLERVQTGIWDIPAGYLPADYFEGVTRA
MLBr_01573|M.leprae_Br4923 --------MDLSGSRPVVGLTAYLEQVHTGLWDVPAGYLPADYFQGVAMA
MMAR_1844|M.marinum_M ------MVLNAAGPGPIVGMTTYLDQAKTGVWDVSASFLPASYFDGVTMA
MUL_2093|M.ulcerans_Agy99 ------MVLNAAGPGPIVGMTTYLDQAKTGVWDVSASFLPASYFDGVTMA
Mflv_4064|M.gilvum_PYR-GCK -------------SRPVIGLTTYLQQAQTGVWDVRASFLPAIYFEGVGMA
Mvan_2279|M.vanbaalenii_PYR-1 QARQRDSVSLRDRTRPVIGLTTYLQQAQTGVWDVRASFLPAIYFEGVGMA
MSMEG_1119|M.smegmatis_MC2_155 -----------MANRPLIGLTTYRESIRWGKWHMPALFTPETYVSAVDRA
:*:*:* : : * *.: * : * *...: *
Mb2884c|M.bovis_AF2122/97 GGVAVLLPPQPVDPESVGCVLDSLHALVITGGYDLDPAAYGQEPHPATDH
Rv2859c|M.tuberculosis_H37Rv GGVAVLLPPQPVDPESVGCVLDSLHALVITGGYDLDPAAYGQEPHPATDH
MAV_3717|M.avium_104 GGIAVLLPPQPVDTGIVGSLLDGLHALVITGGYDLDPAGYGQRPHPSTDA
MLBr_01573|M.leprae_Br4923 GGIAVLLPPQPVDPEIAGLALDGLDGLVITGGYDVDPATYGQRPHPSTDE
MMAR_1844|M.marinum_M GGIAVLLPPQRADADAANRVLDSLDALVITGGKDVDPAAYGQQAHPATDE
MUL_2093|M.ulcerans_Agy99 GGIAVLLPPQRADADAANRVLDSLDALVITGGKDVDPAAYGQQAHPATDE
Mflv_4064|M.gilvum_PYR-GCK GGIASLLPPQPVDDTIAERVLDGIDGLIITGGRDVDPSTYGAQRHPRTDE
Mvan_2279|M.vanbaalenii_PYR-1 GGIAVLLPPQAADAAVADRVLDSLDGLIITGGRDVDPSSYGAQRHPATDE
MSMEG_1119|M.smegmatis_MC2_155 GGEPLLLPTALGHARSA--VLALVDGLLVVGGADIAAENYGERTQPETDP
** . ***. . . * :..*::.** *: . ** . :* **
Mb2884c|M.bovis_AF2122/97 P---RPGRDAWEFALLRGALQRGMPVLGICRGTQVLNVALGGTLHQHLPD
Rv2859c|M.tuberculosis_H37Rv P---RPGRDAWEFALLRGALQRGMPVLGICRGTQVLNVALGGTLHQHLPD
MAV_3717|M.avium_104 P---RTDRDAWEFALLRGALQRGLPVLGICRGAQVLNVAFGGTLHQHLPD
MLBr_01573|M.leprae_Br4923 P---RTTRDSWEFALFEAALQRGLPVLGICRGAQLLNIALGGTLHQHLPE
MMAR_1844|M.marinum_M P---APIRDSWEFALLRGALQRGLPVLGICRGAQVLNVAFGGTLHQHLPD
MUL_2093|M.ulcerans_Agy99 P---APIRDSWEFALLRGALQRGLPVLGICRGAQVLNVAFGGTPHQHLPD
Mflv_4064|M.gilvum_PYR-GCK PDTDSRARDAFEFALVRAALRRAMPVLGICRGAQVLNVALGGTLHQHVPD
Mvan_2279|M.vanbaalenii_PYR-1 PVGDSRTRDAFEFALLQGALRRQIPVLGICRGAQMLNVALGGTLHQHLPD
MSMEG_1119|M.smegmatis_MC2_155 R----PVRDRGELEALDYALGNDLPVLGVCRGMQLINVHAGGSLIQHLPT
** *: . ** . :****:*** *::*: **: **:*
Mb2884c|M.bovis_AF2122/97 ILGHSGHRAGNGVFTRLPVHTASGTRLAELIGESADVPCYHHQAIDQVGE
Rv2859c|M.tuberculosis_H37Rv ILGHSGHRAGNGVFTRLPVHTASGTRLAELIGESADVPCYHHQAIDQVGE
MAV_3717|M.avium_104 VLGHGGHRAGNGVFSTLPVRTVAGTRLAALLGETVDAPCYHHQAIDKLGD
MLBr_01573|M.leprae_Br4923 VIGHSKHWVGNAVFNNLLVRTVPGTRLAAVLGEFVEARCYHHQSIDKLGD
MMAR_1844|M.marinum_M VLGHNGHRAGNAVFSSLPVRTVPGTRLATLIGESAQVRCYHHQAVAEVGE
MUL_2093|M.ulcerans_Agy99 VLGHNGHRAGNAVFSSLPVRTVPGTRLATLIGESAQVRCYHHQAVAEVGE
Mflv_4064|M.gilvum_PYR-GCK VVGHTRHQQGNAVFTTSSIATVPGSRVAALVGPDTEAQCYHHQAIDRLGD
Mvan_2279|M.vanbaalenii_PYR-1 VVGHTRHQQGNAVFTTSSITTVPGTRVAALVGPDIEAQCYHHQAVDRLGD
MSMEG_1119|M.smegmatis_MC2_155 QVGHHEHLRTPGEFGSHPVRVDQASRISTVVGPGPVVTTYHHQGIGRLGS
:** * . * : . .:*:: ::* . ****.: .:*.
Mb2884c|M.bovis_AF2122/97 GLVVSAVDVDGVIEALELP----GDTFVLAVQWHPEKSLDDLRLFKALVD
Rv2859c|M.tuberculosis_H37Rv GLVVSAVDVDGVIEALELP----GDTFVLAVQWHPEKSLDDLRLFKALVD
MAV_3717|M.avium_104 GLVVSAWDPDGVVEAVELP----GDAFVLAVQWHPEQSLHDLRLFTAIVD
MLBr_01573|M.leprae_Br4923 GLVVSAWDADGVVEAVELP----GDAFVLGVQWHPEKALSDLRLFTAIVD
MMAR_1844|M.marinum_M GLVVSAWDVDGVVEALELP----GENFVLAVQWHPEESLEDLRLFSAVVD
MUL_2093|M.ulcerans_Agy99 GLVVSAFDVDGVVEALELP----GENFVLAVQWHPEESLEDLRLFSAVVD
Mflv_4064|M.gilvum_PYR-GCK GLVVSASDTDGVIEAVEVDLSAHPDHWVIAVQWHPEERLDDLRLFAGLVG
Mvan_2279|M.vanbaalenii_PYR-1 GLIVSARGVDGVVEAVELDPATRSDGWAVAVQWHPEERLDDLRLFAGLVS
MSMEG_1119|M.smegmatis_MC2_155 GLKAVAWADDGVIEAVEST----THSYVIGVQWHAE-RDDGTPLIEAFVE
** . * ***:**:* . :.:.****.* . *: ..*
Mb2884c|M.bovis_AF2122/97 AASGYAGRQSQAEPR
Rv2859c|M.tuberculosis_H37Rv AASGYAGRQSQAEPR
MAV_3717|M.avium_104 AARSYAGVAR-----
MLBr_01573|M.leprae_Br4923 AACRYANSRCVEYS-
MMAR_1844|M.marinum_M AARSRRSG-------
MUL_2093|M.ulcerans_Agy99 AARSRRSG-------
Mflv_4064|M.gilvum_PYR-GCK AAGVYATQKTQKVNR
Mvan_2279|M.vanbaalenii_PYR-1 AAGDYAWHKIEKVS-
MSMEG_1119|M.smegmatis_MC2_155 QCARRKAKWP-----
.