For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTETTTTFMDNVLGWLHKGYPEGVPPKDYFALLALLKRSLTEDEVVRAAQAILRSTDGQSPVTDDDIRNA VHQIIEKEPTAEEINQVAARLASVGWPLAVPVR
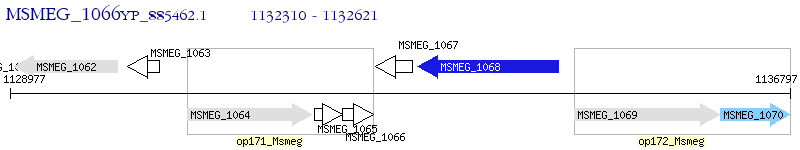
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1066 | - | - | 100% (103) | hypothetical protein MSMEG_1066 |
| M. smegmatis MC2 155 | MSMEG_1063 | - | 6e-13 | 39.36% (94) | hypothetical protein MSMEG_1063 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0557c | - | 4e-16 | 41.76% (91) | hypothetical protein Mb0557c |
| M. gilvum PYR-GCK | Mflv_0029 | - | 3e-30 | 65.22% (92) | hypothetical protein Mflv_0029 |
| M. tuberculosis H37Rv | Rv0543c | - | 4e-16 | 41.76% (91) | hypothetical protein Rv0543c |
| M. leprae Br4923 | MLBr_02258 | - | 1e-13 | 37.36% (91) | hypothetical protein MLBr_02258 |
| M. abscessus ATCC 19977 | MAB_3403c | - | 1e-20 | 45.10% (102) | hypothetical protein MAB_3403c |
| M. marinum M | MMAR_0889 | - | 8e-17 | 43.96% (91) | hypothetical protein MMAR_0889 |
| M. avium 104 | MAV_3912 | - | 9e-14 | 40.59% (101) | hypothetical protein MAV_3912 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0642 | - | 6e-17 | 43.96% (91) | hypothetical protein MUL_0642 |
| M. vanbaalenii PYR-1 | Mvan_0941 | - | 2e-31 | 63.83% (94) | hypothetical protein Mvan_0941 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0889|M.marinum_M MNR--------FLTSIVSWLRAGYPDGVPPTDSFALLALLCRR-LSNEDV
MUL_0642|M.ulcerans_Agy99 MNR--------FLTSIVSWLRAGYPDGVPPTDSFALLALLCRR-LSNEDV
Mb0557c|M.bovis_AF2122/97 MNR--------FLTSIVAWLRAGYPEGIPPTDSFAVLALLCRR-LSHDEV
Rv0543c|M.tuberculosis_H37Rv MNR--------FLTSIVAWLRAGYPEGIPPTDSFAVLALLCRR-LSHDEV
MLBr_02258|M.leprae_Br4923 MNR--------FLTSIVSWLRAGYPEGIPATDTFAVLALLARR-LTNDEV
MAB_3403c|M.abscessus_ATCC_199 MTSTSIIG------SILSWLSAGYPEGVPPTDRFPLLALLRRT-LTEEQV
MAV_3912|M.avium_104 MTNTAASQTVNFCRSVLRWLRAGYPEGVPGPDRVPLLALLRSTPLTEEQI
Mflv_0029|M.gilvum_PYR-GCK MREN-------VFDNVLGWLRHGYPHGIPTTDYFPLLALLSRT-LTEDEV
Mvan_0941|M.vanbaalenii_PYR-1 MAES-------AFNNILDWLHQGYPQGVPQTDYFALLALLART-LSEDEV
MSMEG_1066|M.smegmatis_MC2_155 MTETTTT----FMDNVLGWLHKGYPEGVPPKDYFALLALLKRS-LTEDEV
* .:: ** ***.*:* * ..:**** *:.:::
MMAR_0889|M.marinum_M TTVATELMR--RGEFDQID-------IGVAITQITDDLPSPEDIERVRAR
MUL_0642|M.ulcerans_Agy99 TTVATELMR--RGEFDQID-------IGVAITQITDDLPSPEDIERVRAR
Mb0557c|M.bovis_AF2122/97 KAVANELMR--LGDFDQID-------IGVVITHFTDELPSPEDVERVRAR
Rv0543c|M.tuberculosis_H37Rv KAVANELMR--LGDFDQID-------IGVVITHFTDELPSPEDVERVRAR
MLBr_02258|M.leprae_Br4923 KLVARELIR--RGEFDKID-------IGVMISHLTDELPSPQDIERVRTR
MAB_3403c|M.abscessus_ATCC_199 QEVVAKLTD--PESSAQIDGVVSKDEIEKFIADVTKDEPTAQDISRVASR
MAV_3912|M.avium_104 AEVVREITK--DGSPAIADGVINRDEIAEFISGMTHYDAGPENIIRVAAR
Mflv_0029|M.gilvum_PYR-GCK VGVAQTVLRGTDSD-TVTPQ-----EIRAAIHLVTDKEPNPEEMHQVAAR
Mvan_0941|M.vanbaalenii_PYR-1 VKVAQTVLKGSRSD-AVTPE-----EIRAAIHVVTDKEPTAEEMNQVAAR
MSMEG_1066|M.smegmatis_MC2_155 VRAAQAILRSTDGQSPVTDD-----DIRNAVHQIIEKEPTAEEINQVAAR
.. : . * : . . . .::: :* :*
MMAR_0889|M.marinum_M LAATGWPLDDAREDRA-----------------------
MUL_0642|M.ulcerans_Agy99 LAATGWPLDDAREDRA-----------------------
Mb0557c|M.bovis_AF2122/97 LAAQGWPLDDVRDREEHA---------------------
Rv0543c|M.tuberculosis_H37Rv LAAQGWPLDDVRDREEHA---------------------
MLBr_02258|M.leprae_Br4923 LNAKGWLLDNARDNGEPT---------------------
MAB_3403c|M.abscessus_ATCC_199 LAAGGWPLAGVDSTALNA---------------------
MAV_3912|M.avium_104 LAAAGWPLAGIDVSEVIPGDDFAESAEIAARGQAPGVAS
Mflv_0029|M.gilvum_PYR-GCK LASVGWPLAAPAG--------------------------
Mvan_0941|M.vanbaalenii_PYR-1 LASVGWPLATPTR--------------------------
MSMEG_1066|M.smegmatis_MC2_155 LASVGWPLAVPVR--------------------------
* : ** *