For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NAFLAKIAAWLNAGYPEGVPGPDRVPLLALLTRRLTNDEIKAIAEDLEKRAHFDHIDIGVLITQMTDEMP REEDIERVRRHLALQGWPLDDPRDGEEPDGESGGPR*
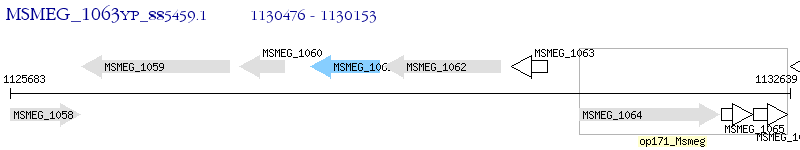
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1063 | - | - | 100% (107) | hypothetical protein MSMEG_1063 |
| M. smegmatis MC2 155 | MSMEG_1066 | - | 6e-13 | 39.36% (94) | hypothetical protein MSMEG_1066 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0557c | - | 7e-32 | 62.24% (98) | hypothetical protein Mb0557c |
| M. gilvum PYR-GCK | Mflv_0032 | - | 2e-32 | 61.54% (104) | hypothetical protein Mflv_0032 |
| M. tuberculosis H37Rv | Rv0543c | - | 7e-32 | 62.24% (98) | hypothetical protein Rv0543c |
| M. leprae Br4923 | MLBr_02258 | - | 2e-30 | 57.58% (99) | hypothetical protein MLBr_02258 |
| M. abscessus ATCC 19977 | MAB_3951 | - | 1e-36 | 68.93% (103) | hypothetical protein MAB_3951 |
| M. marinum M | MMAR_0889 | - | 4e-29 | 58.95% (95) | hypothetical protein MMAR_0889 |
| M. avium 104 | MAV_4601 | - | 3e-22 | 56.25% (80) | hypothetical protein MAV_4601 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0642 | - | 3e-29 | 58.95% (95) | hypothetical protein MUL_0642 |
| M. vanbaalenii PYR-1 | Mvan_0938 | - | 2e-34 | 64.08% (103) | hypothetical protein Mvan_0938 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0557c|M.bovis_AF2122/97 -------------------------------MNRFLTSIVAWLRAGYPEG
Rv0543c|M.tuberculosis_H37Rv -------------------------------MNRFLTSIVAWLRAGYPEG
MMAR_0889|M.marinum_M -------------------------------MNRFLTSIVSWLRAGYPDG
MUL_0642|M.ulcerans_Agy99 -------------------------------MNRFLTSIVSWLRAGYPDG
MLBr_02258|M.leprae_Br4923 -------------------------------MNRFLTSIVSWLRAGYPEG
MAV_4601|M.avium_104 --------------------------------------------------
MAB_3951|M.abscessus_ATCC_1997 MAGDNGQQQHYDRYSVDKIQDGRPSLQELEAVNAFLNKIVTWLRAGYPEG
Mflv_0032|M.gilvum_PYR-GCK -------------------------------MTKFLARIVAWIVEGYPEG
Mvan_0938|M.vanbaalenii_PYR-1 -------------------------------MTRFLARIVAWIAAGYPEG
MSMEG_1063|M.smegmatis_MC2_155 -------------------------------MNAFLAKIAAWLNAGYPEG
Mb0557c|M.bovis_AF2122/97 IPPTDSFAVLALLCRRLSHDEVKAVANELMRLGDFDQ--------IDIGV
Rv0543c|M.tuberculosis_H37Rv IPPTDSFAVLALLCRRLSHDEVKAVANELMRLGDFDQ--------IDIGV
MMAR_0889|M.marinum_M VPPTDSFALLALLCRRLSNEDVTTVATELMRRGEFDQ--------IDIGV
MUL_0642|M.ulcerans_Agy99 VPPTDSFALLALLCRRLSNEDVTTVATELMRRGEFDQ--------IDIGV
MLBr_02258|M.leprae_Br4923 IPATDTFAVLALLARRLTNDEVKLVARELIRRGEFDK--------IDIGV
MAV_4601|M.avium_104 MPPTDTFPVLALLARRLSGDEVKDVASELIRRGEFDD--------VDIGV
MAB_3951|M.abscessus_ATCC_1997 VPPPDYVPLLALLGRRLSNDEVKTVARELMARGDFDN--------IDIGV
Mflv_0032|M.gilvum_PYR-GCK VPGPDRVPLLALLRRRLTDDEVRAVVTELRTRAELTD--TPGIDTVDIGV
Mvan_0938|M.vanbaalenii_PYR-1 IPGADRIPLLALLRRRLTDDEVKAVVAELIARAEFGERGSVDIDPVDIGV
MSMEG_1063|M.smegmatis_MC2_155 VPGPDRVPLLALLTRRLTNDEIKAIAEDLEKRAHFDH--------IDIGV
:* .* ..:**** ***: ::: :. :* ..: . :****
Mb0557c|M.bovis_AF2122/97 VITHFTDELPSPEDVERVRARLAAQGWPLDDVRDREEHA-----------
Rv0543c|M.tuberculosis_H37Rv VITHFTDELPSPEDVERVRARLAAQGWPLDDVRDREEHA-----------
MMAR_0889|M.marinum_M AITQITDDLPSPEDIERVRARLAATGWPLDDAR--EDRA-----------
MUL_0642|M.ulcerans_Agy99 AITQITDDLPSPEDIERVRARLAATGWPLDDAR--EDRA-----------
MLBr_02258|M.leprae_Br4923 MISHLTDELPSPQDIERVRTRLNAKGWLLDNARDNGEPT-----------
MAV_4601|M.avium_104 LITQITDELPTPDDVERVRVRLAAQGWPFDDADQAGDPA-----------
MAB_3951|M.abscessus_ATCC_1997 LITQITDELPTGEDIERIRERLAKKGWPLDDPREPADHGEEPGSGQATDH
Mflv_0032|M.gilvum_PYR-GCK LITEITDGLPSPDDIERVRARLAAQGWPLDDSRDEEDT------------
Mvan_0938|M.vanbaalenii_PYR-1 LITQITDELPSPDDVERVRARLAAQGWPLDDPRDAEEDR-----------
MSMEG_1063|M.smegmatis_MC2_155 LITQMTDEMPREEDIERVRRHLALQGWPLDDPRDGEEPDGESGGPR----
*:.:** :* :*:**:* :* ** :*: :
Mb0557c|M.bovis_AF2122/97 --
Rv0543c|M.tuberculosis_H37Rv --
MMAR_0889|M.marinum_M --
MUL_0642|M.ulcerans_Agy99 --
MLBr_02258|M.leprae_Br4923 --
MAV_4601|M.avium_104 --
MAB_3951|M.abscessus_ATCC_1997 PA
Mflv_0032|M.gilvum_PYR-GCK --
Mvan_0938|M.vanbaalenii_PYR-1 --
MSMEG_1063|M.smegmatis_MC2_155 --