For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGYHGRAAAIRKKSTTTTLRAGAVLGVIMLGLTGCGGQRGESNDAASAEPTGCEITSDARLSIATGNTTG VYYTLGGALAEAISQQTDGKLKATAAETSASLQNIQQLVAGTHQVAFSLADTAADAVNGTASFSEKQPIA ALARLYPNYTQVIARTDAGINSVADMRGKRVSTGAPNSGTEVIANRMLEAAGLGPGDVQTQRTELGKTVE GMKDGSIDALFWSGGLPTSGITDLIVSRRDQVKFIDVTDTLDKLKEINPIYEAGVIPASTYQTPADVSTV VVPNVLLVKDDMDGNTACVLTKTLFERKDDLVKANKAAEDIELQDARETSPVPLHPGAAKALDELGAAN
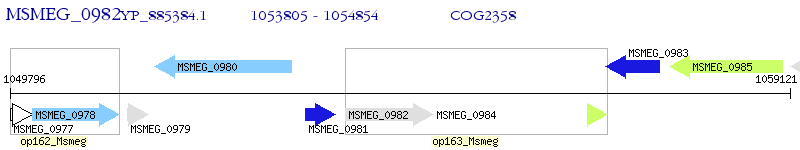
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0982 | - | - | 100% (349) | immunogenic protein |
| M. smegmatis MC2 155 | MSMEG_2634 | - | 7e-05 | 24.38% (201) | hypothetical protein MSMEG_2634 |
| M. smegmatis MC2 155 | MSMEG_0114 | - | 9e-05 | 24.05% (316) | extracellular solute-binding protein, family protein 3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4419 | - | 1e-113 | 63.25% (332) | TRAP transporter solute receptor TAXI family protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_2996 | - | 8e-05 | 25.13% (195) | PUTATIVE putative aliphatic sulfonates family ABC transporter, |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1935 | - | 1e-118 | 64.22% (341) | TRAP transporter solute receptor TAXI family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4419|M.gilvum_PYR-GCK ------------------MTAFATLMLAATAATACGG--RQDAPAADSGG
Mvan_1935|M.vanbaalenii_PYR-1 -----------MKRATQAWAGLAALVLVGAAATGCGG--RQDAPTADSGG
MSMEG_0982|M.smegmatis_MC2_155 MGYHGRAAAIRKKSTTTTLRAGAVLGVIMLGLTGCGGQ-RGESNDAASAE
TH_2996|M.thermoresistible__bu -----------MHTAIEMLRLTLATLISAAALTACGATQSPDSPGLPADA
. : . *.**. :: :
Mflv_4419|M.gilvum_PYR-GCK EITCEVGTETRVSIATGNSTGVYFSLGNAFAEQVSAATDGTVKATAAETG
Mvan_1935|M.vanbaalenii_PYR-1 EITCEVGSDTRVSIATGNSTGVYFSLGNAYAEQISAATDGRVKATAAETG
MSMEG_0982|M.smegmatis_MC2_155 PTGCEITSDARLSIATGNTTGVYYTLGGALAEAISQQTDGKLKATAAETS
TH_2996|M.thermoresistible__bu ALPTEVPPDTTLVVADDANR------LRTLLTLSGEQDRLTGEVSYANFS
*: .:: : :* . . : . :.: *: .
Mflv_4419|M.gilvum_PYR-GCK ASVQNIQQLVAGSYQVAFSLADTASDAIEGTGSFDGKEQPIQALSRIYPN
Mvan_1935|M.vanbaalenii_PYR-1 ASVQNIQQLVGGSYQVAFSLADTAADAVEGKGSFDGEKQPIQAISRIYPN
MSMEG_0982|M.smegmatis_MC2_155 ASLQNIQQLVAGTHQVAFSLADTAADAVNGTASFS-EKQPIAALARLYPN
TH_2996|M.thermoresistible__bu SGPLRLEAIRTGNAHLGY-----VGDVPPILAHYSDADVPIVGAVRRDGT
:. .:: : *. ::.: ..*. . :. . ** . * .
Mflv_4419|M.gilvum_PYR-GCK YTQVIARTDSGITSIADMRGKRVSTGSPGSGTEVIANRLLESAGLNPQTD
Mvan_1935|M.vanbaalenii_PYR-1 YTQVIARTDSGITSIADMRGKRVSTGSPGSGTEVIANRLLQSAGLNPESD
MSMEG_0982|M.smegmatis_MC2_155 YTQVIARTDAGINSVADMRGKRVSTGAPNSGTEVIANRMLEAAGLGP-GD
TH_2996|M.thermoresistible__bu GALIATSPDSGITSLSDLAGKRIGVN-EGTSQQATLLRNLQSVGLGI-GD
: : : .*:**.*::*: ***:... .:. :. * *::.**. *
Mflv_4419|M.gilvum_PYR-GCK VAAQRLDLTKTVDGMKDGSIDAMFWSG--------GLPTPGITDLFTSAR
Mvan_1935|M.vanbaalenii_PYR-1 VAAQRLDLAKTVDGVKDGSIDALFWSG--------GLPTPGITDLLTTSR
MSMEG_0982|M.smegmatis_MC2_155 VQTQRTELGKTVEGMKDGSIDALFWSG--------GLPTSGITDLIVSRR
TH_2996|M.thermoresistible__bu VEPVYLGLAEFADGLRAEQIQAAVLKQPDRARYLESTAAEGAIEIPNAPG
* . * : .:*:: .*:* . . . .: * :: :
Mflv_4419|M.gilvum_PYR-GCK EDVTFIDVTPQLGRMAEISPAYEEGLIPATTYQLP---SDVATIVVPNML
Mvan_1935|M.vanbaalenii_PYR-1 DDVVFIDITPQLPKMSEISPAYEEGVIPAATYQLP---ADVKTIVVPNML
MSMEG_0982|M.smegmatis_MC2_155 DQVKFIDVTDTLDKLKEINPIYEAGVIPASTYQTP---ADVSTVVVPNVL
TH_2996|M.thermoresistible__bu ANTGLNYLYAGRDALIDPARAAAVREFVIAWYRAELWRNGHQDTWISEYL
:. : : : : : : *: . :.: *
Mflv_4419|M.gilvum_PYR-GCK LVRDDLDANLACVLTRT---------------LFEKQPEMAQVVGAAKGI
Mvan_1935|M.vanbaalenii_PYR-1 LVREDLDANLACVLTKT---------------LFEKKPELEQVVGAAKGI
MSMEG_0982|M.smegmatis_MC2_155 LVKDDMDGNTACVLTKT---------------LFERKDDLVKANKAAEDI
TH_2996|M.thermoresistible__bu VKDQHVDVDDARKIVESDGVASIPGFTDEVVATQQETIDLLQQAGAFAGR
: :.:* : * :..: :. :: : * .
Mflv_4419|M.gilvum_PYR-GCK ALDNARDTDPVPLNRGAEYALDELNAPR
Mvan_1935|M.vanbaalenii_PYR-1 NLDNARDTDPVPLNRGAQHALDQLGAAG
MSMEG_0982|M.smegmatis_MC2_155 ELQDARETSPVPLHPGAAKALDELGAAN
TH_2996|M.thermoresistible__bu ELRARDEFDFRFADLTADSPTENRGGQP
* : . . * . :: ..