For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDAAQPGSGPRAERPNRPNRRGLATRENMLEAALQALATGDPGAVSANRIAKQIGATWGAVKYQFGDID GLWAAVLHRTAERRAGVLGRHDPGAPLRQRVAGIIDMLYDGLTASDSRAIENLRAALPRDRAELERLYPR TAAELSSWGQGWLATCQQAFADLDVDPERVREVASFIPGAMRGITSERQLGTYTDLDVARRGLTNAIVSY LEDNGSGRRE
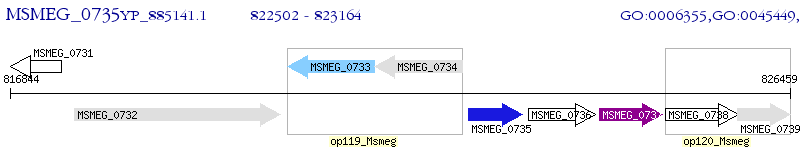
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0735 | - | - | 100% (220) | putative transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_4300 | - | 5e-08 | 29.79% (141) | transcription regulator AmtR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2752 | - | 1e-82 | 76.41% (195) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0725c | - | 3e-78 | 74.23% (194) | hypothetical protein MAB_0725c |
| M. marinum M | MMAR_1934 | - | 2e-81 | 73.13% (201) | hypothetical protein MMAR_1934 |
| M. avium 104 | MAV_3668 | - | 3e-75 | 73.54% (189) | hypothetical protein MAV_3668 |
| M. thermoresistible (build 8) | TH_2354 | - | 1e-105 | 89.20% (213) | putative transcriptional regulator |
| M. ulcerans Agy99 | MUL_3113 | - | 4e-81 | 71.29% (209) | AcrR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_3785 | - | 1e-84 | 74.06% (212) | hypothetical protein Mvan_3785 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0735|M.smegmatis_MC2_155 -------MTDAAQPGSGPRAERPNRPNRRGLATRENMLEAALQALATGDP
TH_2354|M.thermoresistible__bu MPGVSVLKSPGGPPGARP-AEVPRRANRRGQATRENMLEAALAALATGDP
MMAR_1934|M.marinum_M ---------------MTRAGVAGRRPNRRGNATRESMLEAALTSLASGDP
MAV_3668|M.avium_104 ------------------------------------MLEAALRSLASGEP
Mflv_2752|M.gilvum_PYR-GCK ---------------MTEAVA--RRTNRRGQTTRENMLKAALTSLASGDP
Mvan_3785|M.vanbaalenii_PYR-1 ---------------MTEAAAPARRPNRRGQATRANMLEAALKSLASGDP
MAB_0725c|M.abscessus_ATCC_199 ---------------MSEVVATGRRANRRGLATRESMLEAALRVLASGDP
MUL_3113|M.ulcerans_Agy99 ------------MPGMTDLGVQGRRTNRRGHATRESMLEAALRSLASGDP
**:*** **:*:*
MSMEG_0735|M.smegmatis_MC2_155 GAVSANRIAKQIGATWGAVKYQFGDIDGLWAAVLHRTAERRAGVLGRHDP
TH_2354|M.thermoresistible__bu GAVSANRIAKQIGVTWGAVKYQFGDIDGLWAAVLRRTAERRAGVIGRHDA
MMAR_1934|M.marinum_M GSVSANRIAKEIGATWGAVQYQFGDVDGFWAAVLHRTAERRAGSMLALSK
MAV_3668|M.avium_104 GSVSANRIAKDIGATWGAVQYQFGDTDGFWAAVLHRTAERRAATFSSLSS
Mflv_2752|M.gilvum_PYR-GCK GSVSANRIAKDIGATWGAVQYQFGDADGFWAAVLHRTAERRADVFAAVDT
Mvan_3785|M.vanbaalenii_PYR-1 GSVSANRIAKDIGATWGAVQYQFGDADGFWAAVLHRTAERRADVFASVDP
MAB_0725c|M.abscessus_ATCC_199 AAVSANKIAKECGATWGAVKYQFGDIDGLWAAVLQRTAERRGEQPWRNDP
MUL_3113|M.ulcerans_Agy99 GAASANRIAKECGATWGAVKYQFGDIDGFWAAVLHRTAERRGELPSNAVS
.:.***:***: *.*****:***** **:*****:******.
MSMEG_0735|M.smegmatis_MC2_155 ----GAPLRQRVAGIIDMLYDGLTASDSRAIENLRAALPRDRAELERLYP
TH_2354|M.thermoresistible__bu ----GAPLRERVAGIIDMLYDGLTASDSRAIENLRAALPRDRAELERLYP
MMAR_1934|M.marinum_M PLQSGRPLQERVAAVIETLFHGLASADSRAIENLRAALPRDPGELERLYP
MAV_3668|M.avium_104 PVSPEAPLRERVGAIIETLYRGLAAPDSRAIENLRAALPRDPDELERLYP
Mflv_2752|M.gilvum_PYR-GCK ----AAPLRTRVASIIDTLYHGLAEPDSRAIENLRAALPRDHAELERLYP
Mvan_3785|M.vanbaalenii_PYR-1 ----DASLRDRVASIIDTLYDGLAEPDSRAIENLRAALPRDPAELERLYP
MAB_0725c|M.abscessus_ATCC_199 ----VGPLDRRVSAIVETLYRGLTAPDSRAIENLRRALPPEPAELERLYP
MUL_3113|M.ulcerans_Agy99 ----GGPLPERVAAIIDTLYEGLTSTHSRAIETLRRALPNNRDELERNYP
.* **..::: *: **: ..*****.** *** : **** **
MSMEG_0735|M.smegmatis_MC2_155 RTAAELSSWGQGWLATCQQAFADLDVDPERVREVASFIPGAMRGITSERQ
TH_2354|M.thermoresistible__bu RTAAELSSWGEGWLQTCQQAFADLDVDPERVAEVASFIPGAMRGITSERQ
MMAR_1934|M.marinum_M RTAAELFSWGRSWLETCQNAFADLDVDPNRVREVAALIPGAMRGLVSERQ
MAV_3668|M.avium_104 RTAAELFSWGKSWLETCQQAFAGLAVDPDRVREVAALIPGAMRGLVSERQ
Mflv_2752|M.gilvum_PYR-GCK QTAAELHSWGQSWLQICHNAFAGIDVDPDRVREVATLIPGAMRGLVSERQ
Mvan_3785|M.vanbaalenii_PYR-1 QTAAELFSWGQSWLQICHNAFAGLGVDPDRVREVATLIPGAMRGLVSERQ
MAB_0725c|M.abscessus_ATCC_199 RTAAELASWKHRWARACQQAFDGLEVDPVRVREVAAFIPGAMRGLVSEKQ
MUL_3113|M.ulcerans_Agy99 RTAAELSSWQRSWALACQKAFAGLEVDPNRVREVAAFIPGAMRGITSEKQ
:***** ** . * *::** .: *** ** ***::*******:.**:*
MSMEG_0735|M.smegmatis_MC2_155 LGTYTDLDVARRGLTNAIVSYLEDNGSGRRE----
TH_2354|M.thermoresistible__bu LGTYIDLELARRGLTNAIVSYLEDRGPGEKSNQ--
MMAR_1934|M.marinum_M LGSYADLELARRALTNALAGYLKNSRTP-------
MAV_3668|M.avium_104 LGSYADLDMARRGLTNALAAYLEQSRP--------
Mflv_2752|M.gilvum_PYR-GCK LGSYADLDVARRGLTNALVAYLQSPAQPHNDISVT
Mvan_3785|M.vanbaalenii_PYR-1 LGSYADLDVARRGLTNALVAYLQAAD---------
MAB_0725c|M.abscessus_ATCC_199 LGTYSDLEEARRGLTNAIVAYLGGHE---------
MUL_3113|M.ulcerans_Agy99 LGTYSDLDEARRGLTNAIVAYLENSGVR-------
**:* **: ***.****:..**