For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ELPAFPTASDPDDEPVPTGEAFGAPFPGTATEGDPGVGDHTNEIFDLASTASTSENQVPMTVVTNPPETV AVTAYPTGNVARIELSHKVTAMTEAELAEEIVVVADTATKKASALVYGSIVDALVEHGLPRANAQEFAAT NMPFATPEQAKQAELALITRHAEQGG*
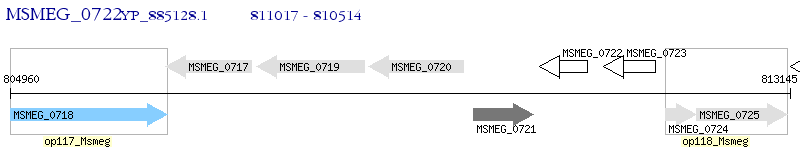
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0722 | - | - | 100% (167) | hypothetical protein MSMEG_0722 |
| M. smegmatis MC2 155 | MSMEG_0058 | - | 3e-18 | 39.71% (136) | hypothetical protein MSMEG_0058 |
| M. smegmatis MC2 155 | MSMEG_0080 | - | 2e-14 | 32.73% (165) | hypothetical protein MSMEG_0080 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3644c | - | 6e-13 | 30.82% (146) | hypothetical protein Mb3644c |
| M. gilvum PYR-GCK | Mflv_0156 | - | 3e-21 | 39.26% (135) | hypothetical protein Mflv_0156 |
| M. tuberculosis H37Rv | Rv3614c | - | 6e-13 | 30.82% (146) | hypothetical protein Rv3614c |
| M. leprae Br4923 | MLBr_00407 | - | 2e-13 | 33.33% (150) | hypothetical protein MLBr_00407 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4350 | - | 2e-13 | 43.62% (94) | hypothetical protein MMAR_4350 |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_2234 | - | 1e-15 | 37.84% (111) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_4592 | - | 1e-13 | 43.62% (94) | hypothetical protein MUL_4592 |
| M. vanbaalenii PYR-1 | Mvan_0071 | - | 2e-19 | 39.39% (132) | hypothetical protein Mvan_0071 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3644c|M.bovis_AF2122/97 ----------------------------MDLPGNDFDSNDFDAVDLWGAD
Rv3614c|M.tuberculosis_H37Rv ----------------------------MDLPGNDFDSNDFDAVDLWGAD
MLBr_00407|M.leprae_Br4923 MGSRRRINRRLLPMSTFPAWQEFRRDVVVVFPGNDFDRDDCDTVDPWGVG
MMAR_4350|M.marinum_M ----------------------------MVQPRSNAHSN-LDALKFSRTA
MUL_4592|M.ulcerans_Agy99 ----------------------------MVQPRSNAHSN-LDALKFSRTA
Mvan_0071|M.vanbaalenii_PYR-1 ----------------------------MANPPPGFEASGFDASGFHDGG
MSMEG_0722|M.smegmatis_MC2_155 ------------------------MELPAFPTASDPDDEP-------VPT
Mflv_0156|M.gilvum_PYR-GCK ------------------------------MPLIDPGDDW-------EDA
TH_2234|M.thermoresistible__bu -----------------------------MTSPYDPDAEWDGESGGVGDS
. . .
Mb3644c|M.bovis_AF2122/97 GAEGWTADPIIGVGSAATP--------DTGPDLDNAHGQAETDTEQ---E
Rv3614c|M.tuberculosis_H37Rv GAEGWTADPIIGVGSAATP--------DTGPDLDNAHGQAETDTEQ---E
MLBr_00407|M.leprae_Br4923 GAAHWTIDPIVGFASSSAPQ-------DRGTDVDNTRGQAEEDEKQKEPE
MMAR_4350|M.marinum_M DNKESAIDALRAFAPTQPE--------EADVDLRALHKPAKDDKAE---E
MUL_4592|M.ulcerans_Agy99 DNKESAIDALRAFAPTQPE--------EADVDLRALHKPAKDDKAE---E
Mvan_0071|M.vanbaalenii_PYR-1 D-----HDDLAALDFSIAGD-------EDEADLGVIFGDFGSASDD-DGE
MSMEG_0722|M.smegmatis_MC2_155 GEAFGAPFPGTATEGD-----------PGVGDHTNEIFDLASTASTSENQ
Mflv_0156|M.gilvum_PYR-GCK GSDLDALV---APEED-----------PGP-------------------E
TH_2234|M.thermoresistible__bu GDDLAAALDFSAPGEDDADVDDLYVAEPEPEEPAEGYVDPYEQAEEPEDL
. .
Mb3644c|M.bovis_AF2122/97 IALFTVTNPPRTVSVSTLMDGRIDHVELSARVAWMSESQLASEILVIADL
Rv3614c|M.tuberculosis_H37Rv IALFTVTNPPRTVSVSTLMDGRIDHVELSARVAWMSESQLASEILVIADL
MLBr_00407|M.leprae_Br4923 VAIFTVTNPPRTVSVSVLMDGRIDHVELSKRVTWMSESQVASEILVLADL
MMAR_4350|M.marinum_M CDLFTVTNPQGTVSVSAVLGGKIRRVELSEEVTTMAEPNLAAEIYVIADL
MUL_4592|M.ulcerans_Agy99 CDLFTVTNPQGTVSVSAVLGGKIRRVELSEEVTTMAEPNLAAEIYVIADL
Mvan_0071|M.vanbaalenii_PYR-1 MPMFTVTNPPGTVTVTTYMDGRVHQIELAPSAAALPEAVLADEIVVIAGL
MSMEG_0722|M.smegmatis_MC2_155 VPMTVVTNPPETVAVTAYPTGNVARIELSHKVTAMTEAELAEEIVVVADT
Mflv_0156|M.gilvum_PYR-GCK IPVVQVTNPSGTITITANLDGSIARVALTPAVTRWTESELAAAITELAEV
TH_2234|M.thermoresistible__bu GPLFSVTNPPGTLTVTAFLDGGIQSVEISPSVTGMTESELAQELLAIAKV
: **** *::::. * : : :: .: .*. :* : :*
Mb3644c|M.bovis_AF2122/97 ARQKAQSAQYAFILDRMSQQVDADEHRVALLRKTVGETWGLPSPEEAAAA
Rv3614c|M.tuberculosis_H37Rv ARQKAQSAQYAFILDRMSQQVDADEHRVALLRKTVGETWGLPSPEEAAAA
MLBr_00407|M.leprae_Br4923 ARQKAQSAQYTFILDKLSQLADGDEHRVALLRESVGNTWNLPSPEQAAEA
MMAR_4350|M.marinum_M ARQKARAAQHTLMVQSMKDIES--EEHRAKLREFLGTMLNLPTPEQAAAA
MUL_4592|M.ulcerans_Agy99 ARQKARAAQHTLMVQSMKDIES--EKHRAKLREFLGTMLNLPTPEQAAAA
Mvan_0071|M.vanbaalenii_PYR-1 ATQDARSAQYTFMLEGMREHGH----DNVTTRDFLNRDLDLPTPEQARAA
MSMEG_0722|M.smegmatis_MC2_155 ATKKASALVYGSIVDALVEHGL----PRANAQEFAATNMPFATPEQAKQA
Mflv_0156|M.gilvum_PYR-GCK AAKRATAVMHVTVVEMFVAQGV----DRTEAREFVSSNMPFATPAQADAA
TH_2234|M.thermoresistible__bu AAMKARAGQYEFLLEATPQEAG----EREFLGEFLQHGVGLPTPEQAAQV
* * : : ::: . :.:* :* .
Mb3644c|M.bovis_AF2122/97 EAEVFATRYSDDCPAPDDES------------------DPW
Rv3614c|M.tuberculosis_H37Rv EAEVFATRYSDDCPAPDDES------------------DPW
MLBr_00407|M.leprae_Br4923 EAEVFATRYSDYCPAQDTEN------------------DQW
MMAR_4350|M.marinum_M QKEVFATRYGRESARKSRGNGLDGVPVFARRVSDVHREADW
MUL_4592|M.ulcerans_Agy99 QKEVFATRYGRESARKSRGNGLDGVPVFARRVSDVHREADW
Mvan_0071|M.vanbaalenii_PYR-1 RAELFTTRYAGDE----------------------------
MSMEG_0722|M.smegmatis_MC2_155 ELALITRHAEQGG----------------------------
Mflv_0156|M.gilvum_PYR-GCK LEELSARYAHEGR----------------------------
TH_2234|M.thermoresistible__bu EAEFVQRYLRDDV----------------------------
.