For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRNICRPGPLPGPRRSPQPAALRPSAELYGFRQDLDARRSHMKPTHLVAASVISVVMSGFFAAPAVATPA EGDVERTDLAEGTTTTPIWIVTAGQPTTLTVQSLLLQPGAGSGWHAHPGPEHSVVNSGSVVLRTAPGCAP VTYGTGQSVFIPAGVPHEVSNQGSEEAEVVVTYTLPANVPARDDAPAMCP
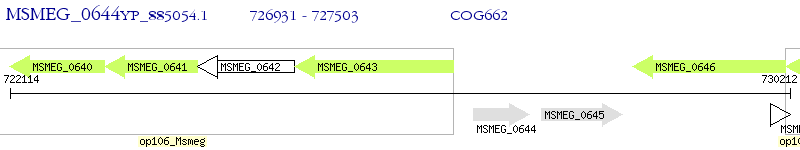
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0644 | - | - | 100% (190) | cupin domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_1754 | - | 4e-07 | 28.09% (89) | hypothetical protein MSMEG_1754 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2547c | - | 4e-06 | 31.78% (107) | hypothetical protein MAB_2547c |
| M. marinum M | MMAR_2152 | - | 4e-05 | 38.46% (78) | transcriptional regulator |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1737 | - | 3e-05 | 38.46% (78) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0588 | - | 2e-47 | 56.79% (162) | cupin 2 domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0644|M.smegmatis_MC2_155 MRNICRPGPLPGPRRSPQPAALRPSAELYGFRQDLDARRSHMKPTHLVAA
Mvan_0588|M.vanbaalenii_PYR-1 ---MIRKANIRRSVR-VEVAASRP----------HTLRGKPMKHVVTVAA
MMAR_2152|M.marinum_M MTALLRAVRRQRGLT-LEQLAQRAGLTKSYLSKIERGQSTPSIAVALKVA
MUL_1737|M.ulcerans_Agy99 MTALLRAVRRQRGLT-LEQLAQRAGLTKSYLSKIERGQSTPSIAVALKVA
MAB_2547c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0644|M.smegmatis_MC2_155 SVISVVMSGFFAAPAVATPAEGDVERTDLAEGTTTTPIWIVTAG--QPTT
Mvan_0588|M.vanbaalenii_PYR-1 SVIAATFA---PATAGATPPEGDVERTELGQGTTNAPVVIVSVG--QPTT
MMAR_2152|M.marinum_M RALDVDVGRLFSDESAHETITVDRAHDRLGPQSGRYHVLASTQLGKTMSP
MUL_1737|M.ulcerans_Agy99 RALDVDVGRLFSDESAHETITADRAHDRLGPQSGRYHVLASTQLGKTMSP
MAB_2547c|M.abscessus_ATCC_199 ------------------MNRHDLQRHDLSTPGR--------------ET
* : *. .
MSMEG_0644|M.smegmatis_MC2_155 LTVQSLLLQPGAGSGWH-AHPGPEHSVVNSGSVVLRTAPGCAPVTYGTGQ
Mvan_0588|M.vanbaalenii_PYR-1 LYVQELELNGPAGSGWH-THPGPEHTVITEGTVHVQSAPDCVPVAYPAGQ
MMAR_2152|M.marinum_M FVVRPTAGQDCASDGPHPEHTGQEFVFVHTGSVELDYG--DQTITLGTGD
MUL_1737|M.ulcerans_Agy99 FVVRPTAGQDCASDGPHPEHTGQEFVFVHTGSVELDYG--DQTITLGTGD
MAB_2547c|M.abscessus_ATCC_199 LQTR-VDFQPGAQAARH-KHPGEEIIYVLKGTLIYDID-GQGSKTVTAGD
: .: : * . * *.* * : *:: . : :*:
MSMEG_0644|M.smegmatis_MC2_155 SVFIPAGVPHEVSNQGSEEAEVVVTYTLPANVPARDDAPAMCP
Mvan_0588|M.vanbaalenii_PYR-1 SVFIPAGVPHTVRNEGPGDAHAVVTYTLPADRAVRDDAPAACP
MMAR_2152|M.marinum_M SAYFDASVGHKLRAVGTERAEVVVVAG--AELGGR--------
MUL_1737|M.ulcerans_Agy99 SAYFDASVGHKLRAVGTERAEVVVVAG--AELGGR--------
MAB_2547c|M.abscessus_ATCC_199 VLFVPAETFHSVRNVGDGEGSELATYVVDKDKPLLVLAT----
:. * . * : * . :.. :