For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VATIAAADRTVTSVRPAELGVKVVIVMALLLIVMLIFAYKVVSLHNMKDEPFWAAYGLIITAFIFTRFGL AALYRPPRRDTGNYWPVVAIIVPAYNEPDIARTLLHCVGVDYPRELLRVVVVDDKSSDDTLRRITDFAAE HPNLTVIPHEVNGGKRRAMATGMAAADDADLFVFIDSDSQVTRDAIRVIASYFADSSVGAVCGHTDVTNI SHNILTRMQAMQYYIAFRIYKSAEALFGSVTCCSGCFSAYRADAVRPVADKWLNQTFLGRPSTFGDDRSL TNYLLRDWRVLYAPDAQAYTNVPEHLKQFLRQQLRWKKSWLREAPRAMCAVRRKNPVMVVMFALSIVLPL IAPQVVMRAFVVQPHFISQLPFWYFGGVAAIAVIYGLFYRLHRPVKRWYQGIFFTMFYTIILVLQMPYAM VTIRDSKWGTR
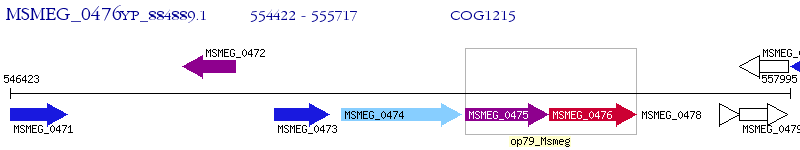
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0476 | - | - | 100% (431) | chitin synthase |
| M. smegmatis MC2 155 | MSMEG_1426 | - | 6e-07 | 24.42% (172) | membrane sugar transferase |
| M. smegmatis MC2 155 | MSMEG_5961 | - | 2e-06 | 28.57% (133) | glycosyl transferase, group 2 family protein, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0715 | - | 8e-07 | 27.74% (155) | membrane sugar transferase |
| M. gilvum PYR-GCK | Mflv_0207 | - | 8e-10 | 24.70% (247) | hypothetical protein Mflv_0207 |
| M. tuberculosis H37Rv | Rv0696 | - | 8e-07 | 27.74% (155) | membrane sugar transferase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3831c | - | 7e-07 | 23.28% (189) | putative glycosyltransferase |
| M. marinum M | MMAR_1024 | - | 2e-08 | 28.10% (153) | membrane glycosyl transferase |
| M. avium 104 | MAV_4475 | - | 1e-07 | 28.95% (152) | membrane sugar transferase |
| M. thermoresistible (build 8) | TH_1903 | - | 5e-06 | 25.60% (125) | PROBABLE MEMBRANE SUGAR TRANSFERASE |
| M. ulcerans Agy99 | MUL_0776 | - | 4e-08 | 32.08% (106) | membrane glycosyl transferase |
| M. vanbaalenii PYR-1 | Mvan_1299 | - | 1e-07 | 28.80% (125) | glycosyl transferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
TH_1903|M.thermoresistible__bu ------------MTGQRLPDGFAVQ-VDRRVKVLGEGAALLGGSPTRLLK
Mvan_1299|M.vanbaalenii_PYR-1 ------------MTGPRLPDGFAVQ-VDRRVRVLGEGAALLGGSPTRLLR
Mb0715|M.bovis_AF2122/97 ------------MTATRLPDGFAVQ-VDRRVRVLGDGSALLGGSPTRLLR
Rv0696|M.tuberculosis_H37Rv ------------MTATRLPDGFAVQ-VDRRVRVLGDGSALLGGSPTRLLR
MMAR_1024|M.marinum_M ------------MTQPRLPDGFAVQ-VDRRVRVLGGGSALLGGSPTRLLR
MUL_0776|M.ulcerans_Agy99 ------------MTQPRLPDGFAVQ-VDRRVRVLGGGSALLGGSPTRLLR
MAV_4475|M.avium_104 ----------MTAPAPRLPDGFAVQ-VDRRVRVLGDGSALLGGSPTRLLK
MAB_3831c|M.abscessus_ATCC_199 MTTSDEQAQHAAPSQERLPDGFAVQ-VDRRVRVLDEGSALLGGSPTRLLR
Mflv_0207|M.gilvum_PYR-GCK --------MHSYTPGSRLPEDLSARRVTTDGSVILGVSGILGVAATGVLA
MSMEG_0476|M.smegmatis_MC2_155 ----------MATIAAADRTVTSVRPAELGVKVVIVMALLLIVMLIFAYK
:.: . *: : :*
TH_1903|M.thermoresistible__bu ---LAPAARTMLAGGRLEVRDAVSAQLAR-----TLLDATVAHPRPASGP
Mvan_1299|M.vanbaalenii_PYR-1 ---LAPAAQTMLHGGRLEVHDAVSAQLAR-----TLLDATVAHPRPMSGP
Mb0715|M.bovis_AF2122/97 ---LAPAARGLLCDGRLKVRDEVSAELAR-----ILLDATVAHPRPPSGP
Rv0696|M.tuberculosis_H37Rv ---LAPAARGLLCDGRLKVRDEVSAELAR-----ILLDATVAHPRPPSGP
MMAR_1024|M.marinum_M ---LAPAAQDMLCDGRLKVRDDISAELAR-----TLLDATVAHPRPASGP
MUL_0776|M.ulcerans_Agy99 ---LAPAAQDMLCDGRLKVRDDISAELAR-----TLLDATVAHPRPASGP
MAV_4475|M.avium_104 ---LAPAAQDLLCDGRLKVRDDVSAQLAR-----TLLDATVAHPRPAGGP
MAB_3831c|M.abscessus_ATCC_199 ---LAPAARTLLSGGRLEVRDATSAQLAR-----TLLDATVAHPRPVGGP
Mflv_0207|M.gilvum_PYR-GCK PHLVLPAVTTVLAALYLASTIDRHWLLVQGLRSPSLLTISDEEARAVPDN
MSMEG_0476|M.smegmatis_MC2_155 VVSLHNMKDEPFWAAYGLIITAFIFTRFG-------LAALYRPPRRDTGN
: : * .* .
TH_1903|M.thermoresistible__bu SHRDVTVVIPVRDNAFGLHRLMRALRGLR--------VIIVDD---GSAV
Mvan_1299|M.vanbaalenii_PYR-1 SHRDVTVIIPVRDNLSGLTRLVSALRGLR--------VVVVDD---GSAV
Mb0715|M.bovis_AF2122/97 SHRDVTVVIPVRNNASGLRRLVTSLRGLR--------VIVVDD---GSAC
Rv0696|M.tuberculosis_H37Rv SHRDVTVVIPVRNNASGLRRLVTSLRGLR--------VIVVDD---GSAC
MMAR_1024|M.marinum_M SHRDVTVVIPVRDNASGVRRLVASLRGLR--------VIVVDD---GSSS
MUL_0776|M.ulcerans_Agy99 SHRDVTVVISVRDNASGVRRLVASLRGLR--------VIVVDD---GSSS
MAV_4475|M.avium_104 SHHDVTVVIPVRDNLSGVRRLVSSLRGLR--------VVVVDD---GSFP
MAB_3831c|M.abscessus_ATCC_199 GYRDVTVVIPCRDNGFGLRRLLRALRGMR--------VIVVDD---GSTI
Mflv_0207|M.gilvum_PYR-GCK QLPVYTVLLPVYNEPSIVHNLIAGVGRLEYPKDKLEILLLVEEDDIATRR
MSMEG_0476|M.smegmatis_MC2_155 YWPVVAIIVPAYNEPDIARTLLHCVGVDYPR--ELLRVVVVDDKS-SDDT
::::. :: *: : :::*:: .
TH_1903|M.thermoresistible__bu PVDAAEFTGLHCDVQVLRHPHSKGPAAARNTGLAACT-TDFVAFLDSDVV
Mvan_1299|M.vanbaalenii_PYR-1 PVAESDFAATRCDVQVLRNDRSKGPAAARNAGLAVCT-TDLVAFLDSDVL
Mb0715|M.bovis_AF2122/97 PVESDDFVGAHCDIEVLHHPHSKGPAAARNTGLAACT-TDFVAFLDSDVT
Rv0696|M.tuberculosis_H37Rv PVESDDFVGAHCDIEVLHHPHSKGPAAARNTGLAACT-TDFVAFLDSDVT
MMAR_1024|M.marinum_M PVELEDFAGAHCDVEVLHHRRSKGPAAARNTGLAACT-TEFVAFLDSDVA
MUL_0776|M.ulcerans_Agy99 PVELEDFAGAHCDIEVLHHRRSKGPAAARNTGLAACT-TEFVAFLDSDVA
MAV_4475|M.avium_104 PIEPEDFVGAHCDIEVLRHHRSKGPAAARNTGLAACR-TDFVAFLDSDVA
MAB_3831c|M.abscessus_ATCC_199 PIVESDLEGMHCHVEVVRHADSQGPAAARNTGLKLAT-TDFVAFLDSDVV
Mflv_0207|M.gilvum_PYR-GCK AMATTELEAVRLILVPNSQPKTKPKACNYGMATPGLK-GEMVTIYDAEDI
MSMEG_0476|M.smegmatis_MC2_155 LRRITDFAAEHPNLTVIPHEVNGGKRRAMATGMAAADDADLFVFIDSDSQ
:: . : : : . . ::..: *::
TH_1903|M.thermoresistible__bu PQRGWLEALLGHFCDP--AVALVAPRIVGLRKDDNLIARYEAVRSSLDLG
Mvan_1299|M.vanbaalenii_PYR-1 PRRGWLEALLGHFCDP--AVALVAPRIVALNQSDNVVARYEAVRSSLDLG
Mb0715|M.bovis_AF2122/97 PRRGWLESLLGHFCDP--TVALVAPRIVSLVEGENPVARYEALHSSLDLG
Rv0696|M.tuberculosis_H37Rv PRRGWLESLLGHFCDP--TVALVAPRIVSLVEGENPVARYEALHSSLDLG
MMAR_1024|M.marinum_M PRRGWLEALLGHFCDP--TVALVAPRIVGLAPSENLVARYEAVRSSLDLG
MUL_0776|M.ulcerans_Agy99 PRRGWLEALLGHFCDP--TVALVAPRIVGLAPSENLVARYEAVRSSLDLG
MAV_4475|M.avium_104 PRRGWLEALLGHFCDP--TVGLVAPRIVGLSHSENVVARYEAVHSSLDLG
MAB_3831c|M.abscessus_ATCC_199 PRRGWLEALLGHFSDP--AVALVAPRIVGLVLSDNAIARYEAVRSSLDLG
Mflv_0207|M.gilvum_PYR-GCK PDPLQLRKTVVAFQQLPDNVGCIQARLGYFNEEQNLLTRWFSMEYDQWFG
MSMEG_0476|M.smegmatis_MC2_155 VTRDAIRVIASYFADS--SVGAVCGHTDVTNISHNILTRMQAMQYYIAFR
:. * : *. : : .* ::* ::. :
TH_1903|M.thermoresistible__bu VREAPVVPYGPVSYVPSAAIICRRKVLDEIGGFDEALRSGEDVDLCWRLV
Mvan_1299|M.vanbaalenii_PYR-1 LREAPVIPYGTVSYVPSAAIICRRSRLLEVGGFDESLISGEDVDLCWRLN
Mb0715|M.bovis_AF2122/97 QREAPVLPHSTVSYVPSAAIVCRSSAIRDVGGFDETMHSGEDVDLCWRLI
Rv0696|M.tuberculosis_H37Rv QREAPVLPHSTVSYVPSAAIVCRSSAIRDVGGFDETMHSGEDVDLCWRLI
MMAR_1024|M.marinum_M QREAPILPHSPVSYVPSAAIICRCSAIRQVGGFDETMHSGEDVDLCWRLI
MUL_0776|M.ulcerans_Agy99 QREAPILPHSPVSYVPSAAIICRCSAIRQVGGFDETMHSGEDVDLCWRLI
MAV_4475|M.avium_104 EREAPVLPHSTVSYVPSAAIICRCSAIREIGGFDETLQSGEDVDLCWRLI
MAB_3831c|M.abscessus_ATCC_199 LREAPVVPYGPVSYVPSAAIVVRRSAINEIGGFDEALQCGEDVDLCWRLI
Mflv_0207|M.gilvum_PYR-GCK MTLPAVEAAGCVVPLGGTSNHMRTSVWRAIGGWDEFN-VTEDADLGVRLA
MSMEG_0476|M.smegmatis_MC2_155 IYKSAEALFGSVTCCSGCFSAYRADAVRPVA----------DKWLNQTFL
.. . * . * . :. * * :
TH_1903|M.thermoresistible__bu EAGTRLRYEPIALVAHDHRTDIGEWLGRKAFYGSSAAPLTARHPDKTAPL
Mvan_1299|M.vanbaalenii_PYR-1 EAGARLRYEPIAMVAHDHRTELRKWFARKSFYGSSAAPLTIRHPGKTAPL
Mb0715|M.bovis_AF2122/97 EAGARLRYEPIALVAHDHRTQLRDWIARKAFYGGSAAPLAVRHPDKTAPL
Rv0696|M.tuberculosis_H37Rv EAGARLRYEPIALVAHDHRTQLRDWIARKAFYGGSAAPLAVRHPDKTAPL
MMAR_1024|M.marinum_M ESGARLRYEPIALVGHEHRTQLRAWIARKAFYGGSAAPLSVRHPDKTAPL
MUL_0776|M.ulcerans_Agy99 ESGARLRYEPIALVGHEHRTQLRAWIARKAFYGGSAAPLSVRHPDKTAPL
MAV_4475|M.avium_104 EAGVRLRYEPIALVGHDHRTELRDWLARKAFYGGSAAPLSVRHPDKTAPV
MAB_3831c|M.abscessus_ATCC_199 EAGSRLRYEPVSHVAHDHRLTLREWFARKAFYGKSAAPLSTRHPDKVAPM
Mflv_0207|M.gilvum_PYR-GCK RAGYRTRILDSVTLEEANSDVLNWIRQRSRWYKGYLQTMLVHLRHPAALW
MSMEG_0476|M.smegmatis_MC2_155 GR--------PSTFGDDRSLTNYLLRDWRVLYAPDAQAYTNVPEHLKQFL
. . . * .
TH_1903|M.thermoresistible__bu LMSGWTLVLGVLVAMGSWVGYLAALVLAVITGRRVAKSLSTVGSEPVEVV
Mvan_1299|M.vanbaalenii_PYR-1 VISGWTLVVWMLVAIGSGIGYFASIAVAAITGRRIAKSLASVQTEPMEVA
Mb0715|M.bovis_AF2122/97 VISGGALMAWILMSIGTGLGRLASLVIAVLTGRRIARAMRCAETSFLDVL
Rv0696|M.tuberculosis_H37Rv VISGGALMAWILMSIGTGLGRLASLVIAVLTGRRIARAMRCAETSFLDVL
MMAR_1024|M.marinum_M VISGWALTAWVLMSLGTRATQLASLIIAVLTGRRIATALRSAQTSSWDVV
MUL_0776|M.ulcerans_Agy99 VISGWALTAWVLLSLGTRATQLASLIIAVLTGRRIATALRSAQTSSWDVV
MAV_4475|M.avium_104 VISGWALMTWILMAFGSTLSRLASLLLAVLTGRRIARAMRSAETSMTDVV
MAB_3831c|M.abscessus_ATCC_199 VISRWTLLVWILAAVGSGMGYLAAVGMAALAAGRVARTLRGVDTPPRDVV
Mflv_0207|M.gilvum_PYR-GCK SQVGGKGILRLLNMTG-------AVPIVAVINLVFWATMAAWVLGRPDVV
MSMEG_0476|M.smegmatis_MC2_155 RQQLRWKKSWLREAPR-------AMCAVRRKNPVMVVMFALSIVLPLIAP
: :: . . : .
TH_1903|M.thermoresistible__bu ALAAQGLGSAARQLAGAICRHYWPIALIAALLSPRFRQVVVVAAVVDGVV
Mvan_1299|M.vanbaalenii_PYR-1 VVAAHGLGSAALQLASAICRHYWPIALIAALVSRRSRRVVVVAAVLDGVF
Mb0715|M.bovis_AF2122/97 AVATRGLWAAALQLASAICRHYWPLALLAAILSRRCRRVVLIAAVVDGVV
Rv0696|M.tuberculosis_H37Rv AVATRGLWAAALQLASAICRHYWPLALLAAILSRRCRRVVLIAAVVDGVV
MMAR_1024|M.marinum_M VVAARGLWSAALQLASAICRHYWPVTLIAVIVSRHCRRVVLVAAVIDGVV
MUL_0776|M.ulcerans_Agy99 VVAARGLWSAALQLASAICRHYWPVTLIAVIVSRHCRRVVLVAAVIDGVV
MAV_4475|M.avium_104 TVAGRGLWSAALQLASALCRHYWPLALLAATMSRHFRRVVLVAAVMDGVV
MAB_3831c|M.abscessus_ATCC_199 RVAAQGVGGAALQIASALCRHYWPLALFAAALSRRSRQVLVVAAIVDGVV
Mflv_0207|M.gilvum_PYR-GCK ELAFPGATYYVYLTMYVVGAPLS--VFMGLIVTQRLGKPYMWWAAALVPL
MSMEG_0476|M.smegmatis_MC2_155 QVVMRAFVVQPHFISQLPFWYFGGVAAIAVIYG-------LFYRLHRPVK
:. . . :. :
TH_1903|M.thermoresistible__bu DWLSRREHAGEDTKPVGLFAYLVLKRLDDLAYGCGLWAGVLRERDLGALK
Mvan_1299|M.vanbaalenii_PYR-1 DWVTRNGNADEDTKRVGLPTYLLLKRLDDIAYGLGLWTGVVRERHAGALK
Mb0715|M.bovis_AF2122/97 DWLRRREGADDDAEPIGPLTYLVLKRVDDLAYGAGLWYGVVRERNIGALK
Rv0696|M.tuberculosis_H37Rv DWLRRREGADDDAEPIGPLTYLVLKRVDDLAYGAGLWYGVVRERNIGALK
MMAR_1024|M.marinum_M DWLRRREDPDGDSEPIGLLTYLLLKRIDDLAYGLGLWYGVMRERNAGALK
MUL_0776|M.ulcerans_Agy99 DWLRRREDPDGDSEPIGLLTYLLLKRIDDLAYGLGLWYGVMRERNAGALK
MAV_4475|M.avium_104 DWLRRRDAVGDDVEPIGLPTYLVLKRVDDLAYGLGLWWGVLRERNARALR
MAB_3831c|M.abscessus_ATCC_199 DWMKRNDSSASPDDRIGLIEYVLLKRLDDIAYGIGLWSGIVRERDLGALR
Mflv_0207|M.gilvum_PYR-GCK YWMLQSIAALKAVFQLVTRPQFWEKTVHGLSDTVDVPNSTGRPT------
MSMEG_0476|M.smegmatis_MC2_155 RWYQGIFFTMFYTIILVLQMPYAMVTIRDSKWGTR---------------
* : : .
TH_1903|M.thermoresistible__bu PQIRI
Mvan_1299|M.vanbaalenii_PYR-1 PQIRT
Mb0715|M.bovis_AF2122/97 PQIRT
Rv0696|M.tuberculosis_H37Rv PQIRT
MMAR_1024|M.marinum_M PQIRR
MUL_0776|M.ulcerans_Agy99 PQIRR
MAV_4475|M.avium_104 PQIRS
MAB_3831c|M.abscessus_ATCC_199 PELRP
Mflv_0207|M.gilvum_PYR-GCK -----
MSMEG_0476|M.smegmatis_MC2_155 -----