For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTGPRLPDGFAVQVDRRVKVLGEGAALLGGSPTRLLRLAPTAQNMLSGGRLEVHDAVSAQLARTLLDATV AHPRPASGPSHLDVTVVVPVRDNASGLHRLMAALRGLRVIVVDDGSAIPVQPSDFSGMHCDVQVLRHTRS NGPAAARNTGLASCETDFVAFLDSDVVPKRGWLEALLGHFCDPAVALVAPRIVGLHNADNIVARYESVRS SLDLGVREAPVVPHGTVSYVPSAAIICRRSALVEVGGFDETMHSGEDVDLCWRLVESGARLRYEPIALVA HDHRTNLRAWFHRKAFYGTSAAPLTVRHPGKTSPLVISGWTLMVWLMLGVGSFFGYLASLAAAVFAGTRI ARALSVVETEPKEVAVVAAHGLWSSALQLCSAICRHYWPIAMIAAVLFRRARHAVLVAAVVDGVVDWVTR RGNADDDTKPVGLLTHIVLKRLDDIAYGTGLWTGVVRERHLGALKPQVRS
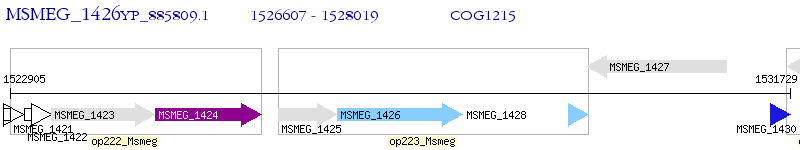
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1426 | - | - | 100% (470) | membrane sugar transferase |
| M. smegmatis MC2 155 | MSMEG_0476 | - | 6e-07 | 24.42% (172) | chitin synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0715 | - | 0.0 | 72.77% (470) | membrane sugar transferase |
| M. gilvum PYR-GCK | Mflv_5057 | - | 0.0 | 77.66% (470) | glycosyl transferase family protein |
| M. tuberculosis H37Rv | Rv0696 | - | 0.0 | 72.77% (470) | membrane sugar transferase |
| M. leprae Br4923 | MLBr_00752 | wbbL | 3e-05 | 37.93% (87) | putative dTDP-rhamnosyl transferase |
| M. abscessus ATCC 19977 | MAB_3831c | - | 0.0 | 68.82% (465) | putative glycosyltransferase |
| M. marinum M | MMAR_1024 | - | 0.0 | 73.56% (469) | membrane glycosyl transferase |
| M. avium 104 | MAV_4475 | - | 0.0 | 71.73% (467) | membrane sugar transferase |
| M. thermoresistible (build 8) | TH_1903 | - | 0.0 | 76.55% (469) | PROBABLE MEMBRANE SUGAR TRANSFERASE |
| M. ulcerans Agy99 | MUL_0776 | - | 0.0 | 73.35% (469) | membrane glycosyl transferase |
| M. vanbaalenii PYR-1 | Mvan_1299 | - | 0.0 | 79.15% (470) | glycosyl transferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0715|M.bovis_AF2122/97 --MTA------------TRLPDGFAVQVDRRVRVLGDGSALLGGSPTRLL
Rv0696|M.tuberculosis_H37Rv --MTA------------TRLPDGFAVQVDRRVRVLGDGSALLGGSPTRLL
MMAR_1024|M.marinum_M --MTQ------------PRLPDGFAVQVDRRVRVLGGGSALLGGSPTRLL
MUL_0776|M.ulcerans_Agy99 --MTQ------------PRLPDGFAVQVDRRVRVLGGGSALLGGSPTRLL
MAV_4475|M.avium_104 MTAPA------------PRLPDGFAVQVDRRVRVLGDGSALLGGSPTRLL
Mflv_5057|M.gilvum_PYR-GCK --MTG------------PRLPDGFAVQVDRRVRVLGEGAALLGGSPTRLL
Mvan_1299|M.vanbaalenii_PYR-1 --MTG------------PRLPDGFAVQVDRRVRVLGEGAALLGGSPTRLL
MSMEG_1426|M.smegmatis_MC2_155 --MTG------------PRLPDGFAVQVDRRVKVLGEGAALLGGSPTRLL
TH_1903|M.thermoresistible__bu --MTG------------QRLPDGFAVQVDRRVKVLGEGAALLGGSPTRLL
MAB_3831c|M.abscessus_ATCC_199 --MTTSDEQAQHAAPSQERLPDGFAVQVDRRVRVLDEGSALLGGSPTRLL
MLBr_00752|M.leprae_Br4923 -------------------MTDVLPVVAVT-------------------Y
:.* :.* .
Mb0715|M.bovis_AF2122/97 RLAPAARGLLCDGRLKVRDEVSAELARILLDATVAHPRPPSGPSHRDVTV
Rv0696|M.tuberculosis_H37Rv RLAPAARGLLCDGRLKVRDEVSAELARILLDATVAHPRPPSGPSHRDVTV
MMAR_1024|M.marinum_M RLAPAAQDMLCDGRLKVRDDISAELARTLLDATVAHPRPASGPSHRDVTV
MUL_0776|M.ulcerans_Agy99 RLAPAAQDMLCDGRLKVRDDISAELARTLLDATVAHPRPASGPSHRDVTV
MAV_4475|M.avium_104 KLAPAAQDLLCDGRLKVRDDVSAQLARTLLDATVAHPRPAGGPSHHDVTV
Mflv_5057|M.gilvum_PYR-GCK RLAPAAQTMLHGGRLEVHDAVSAQLARTLLDATVAHPRPLSGPSHRDVTV
Mvan_1299|M.vanbaalenii_PYR-1 RLAPAAQTMLHGGRLEVHDAVSAQLARTLLDATVAHPRPMSGPSHRDVTV
MSMEG_1426|M.smegmatis_MC2_155 RLAPTAQNMLSGGRLEVHDAVSAQLARTLLDATVAHPRPASGPSHLDVTV
TH_1903|M.thermoresistible__bu KLAPAARTMLAGGRLEVRDAVSAQLARTLLDATVAHPRPASGPSHRDVTV
MAB_3831c|M.abscessus_ATCC_199 RLAPAARTLLSGGRLEVRDATSAQLARTLLDATVAHPRPVGGPGYRDVTV
MLBr_00752|M.leprae_Br4923 SPGPHLERFLASLALATDRPVSVVLAD---NGSTDGTPQVAVERYVNVRL
.* . :* . * . *. ** :.:. . . : :* :
Mb0715|M.bovis_AF2122/97 VIPVRNNASGLRRLVTSLRGLRVIVVDDGSACPVESDDFVGAHCDIEVLH
Rv0696|M.tuberculosis_H37Rv VIPVRNNASGLRRLVTSLRGLRVIVVDDGSACPVESDDFVGAHCDIEVLH
MMAR_1024|M.marinum_M VIPVRDNASGVRRLVASLRGLRVIVVDDGSSSPVELEDFAGAHCDVEVLH
MUL_0776|M.ulcerans_Agy99 VISVRDNASGVRRLVASLRGLRVIVVDDGSSSPVELEDFAGAHCDIEVLH
MAV_4475|M.avium_104 VIPVRDNLSGVRRLVSSLRGLRVVVVDDGSFPPIEPEDFVGAHCDIEVLR
Mflv_5057|M.gilvum_PYR-GCK VIPVHNNATGLTRLVAALRGLKVVIVDDGSSRPVNEADFASAACDIRVLR
Mvan_1299|M.vanbaalenii_PYR-1 IIPVRDNLSGLTRLVSALRGLRVVVVDDGSAVPVAESDFAATRCDVQVLR
MSMEG_1426|M.smegmatis_MC2_155 VVPVRDNASGLHRLMAALRGLRVIVVDDGSAIPVQPSDFSGMHCDVQVLR
TH_1903|M.thermoresistible__bu VIPVRDNAFGLHRLMRALRGLRVIIVDDGSAVPVDAAEFTGLHCDVQVLR
MAB_3831c|M.abscessus_ATCC_199 VIPCRDNGFGLRRLLRALRGMRVIVVDDGSTIPIVESDLEGMHCHVEVVR
MLBr_00752|M.leprae_Br4923 FETGANLGYGT---AANLAIAQFFEEGGATGQPWVDDWVIVANPDVQWGP
. . : * * :.. ...: * . .:.
Mb0715|M.bovis_AF2122/97 HPHSKGPAAARNTGLAACTTDFVAFLDSDVTPRRGWLESLLGHFCDPTVA
Rv0696|M.tuberculosis_H37Rv HPHSKGPAAARNTGLAACTTDFVAFLDSDVTPRRGWLESLLGHFCDPTVA
MMAR_1024|M.marinum_M HRRSKGPAAARNTGLAACTTEFVAFLDSDVAPRRGWLEALLGHFCDPTVA
MUL_0776|M.ulcerans_Agy99 HRRSKGPAAARNTGLAACTTEFVAFLDSDVAPRRGWLEALLGHFCDPTVA
MAV_4475|M.avium_104 HHRSKGPAAARNTGLAACRTDFVAFLDSDVAPRRGWLEALLGHFCDPTVG
Mflv_5057|M.gilvum_PYR-GCK HSRRKGPSAARNAGLAVCATDFVAFLDSDVVPRKGWLEALLGHFCDPAVA
Mvan_1299|M.vanbaalenii_PYR-1 NDRSKGPAAARNAGLAVCTTDLVAFLDSDVLPRRGWLEALLGHFCDPAVA
MSMEG_1426|M.smegmatis_MC2_155 HTRSNGPAAARNTGLASCETDFVAFLDSDVVPKRGWLEALLGHFCDPAVA
TH_1903|M.thermoresistible__bu HPHSKGPAAARNTGLAACTTDFVAFLDSDVVPQRGWLEALLGHFCDPAVA
MAB_3831c|M.abscessus_ATCC_199 HADSQGPAAARNTGLKLATTDFVAFLDSDVVPRRGWLEALLGHFSDPAVA
MLBr_00752|M.leprae_Br4923 GSIDALLDAAARWPRAGALGPLIRDPDGSVYPSARHLPSLVRGGMHAVLG
** . . :: *..* * * :*: ...:.
Mb0715|M.bovis_AF2122/97 LVAPRIVSLVEGENPVARYEALHSSLDLGQREAPVLPHSTVSYVPSAAIV
Rv0696|M.tuberculosis_H37Rv LVAPRIVSLVEGENPVARYEALHSSLDLGQREAPVLPHSTVSYVPSAAIV
MMAR_1024|M.marinum_M LVAPRIVGLAPSENLVARYEAVRSSLDLGQREAPILPHSPVSYVPSAAII
MUL_0776|M.ulcerans_Agy99 LVAPRIVGLAPSENLVARYEAVRSSLDLGQREAPILPHSPVSYVPSAAII
MAV_4475|M.avium_104 LVAPRIVGLSHSENVVARYEAVHSSLDLGEREAPVLPHSTVSYVPSAAII
Mflv_5057|M.gilvum_PYR-GCK LVAPRIVAHEPSDNVVARYEAVRSSLDLGLREAPVIPFGTVSYVPSAAII
Mvan_1299|M.vanbaalenii_PYR-1 LVAPRIVALNQSDNVVARYEAVRSSLDLGLREAPVIPYGTVSYVPSAAII
MSMEG_1426|M.smegmatis_MC2_155 LVAPRIVGLHNADNIVARYESVRSSLDLGVREAPVVPHGTVSYVPSAAII
TH_1903|M.thermoresistible__bu LVAPRIVGLRKDDNLIARYEAVRSSLDLGVREAPVVPYGPVSYVPSAAII
MAB_3831c|M.abscessus_ATCC_199 LVAPRIVGLVLSDNAIARYEAVRSSLDLGLREAPVVPYGPVSYVPSAAIV
MLBr_00752|M.leprae_Br4923 PVWPRNP-------WTAAYRQER--LELTER--------PVGWLSGSCLL
* ** * *. : *:* * .*.::..:.::
Mb0715|M.bovis_AF2122/97 CRSSAIRDVGGFDETMHS-GEDVDLCWRLIEAGARLRYEPIALVAHDHRT
Rv0696|M.tuberculosis_H37Rv CRSSAIRDVGGFDETMHS-GEDVDLCWRLIEAGARLRYEPIALVAHDHRT
MMAR_1024|M.marinum_M CRCSAIRQVGGFDETMHS-GEDVDLCWRLIESGARLRYEPIALVGHEHRT
MUL_0776|M.ulcerans_Agy99 CRCSAIRQVGGFDETMHS-GEDVDLCWRLIESGARLRYEPIALVGHEHRT
MAV_4475|M.avium_104 CRCSAIREIGGFDETLQS-GEDVDLCWRLIEAGVRLRYEPIALVGHDHRT
Mflv_5057|M.gilvum_PYR-GCK CRRSAILEIGGFDETLVS-GEDVDLCWRLNESGARLRYEPIAMVGHDHRT
Mvan_1299|M.vanbaalenii_PYR-1 CRRSRLLEVGGFDESLIS-GEDVDLCWRLNEAGARLRYEPIAMVAHDHRT
MSMEG_1426|M.smegmatis_MC2_155 CRRSALVEVGGFDETMHS-GEDVDLCWRLVESGARLRYEPIALVAHDHRT
TH_1903|M.thermoresistible__bu CRRKVLDEIGGFDEALRS-GEDVDLCWRLVEAGTRLRYEPIALVAHDHRT
MAB_3831c|M.abscessus_ATCC_199 VRRSAINEIGGFDEALQC-GEDVDLCWRLIEAGSRLRYEPVSHVAHDHRL
MLBr_00752|M.leprae_Br4923 VRRSAFREVGGFDERYFMYMEDVDLGDRLGKAGWLSVYVPSAEVLHHKG-
* . : ::***** ***** ** ::* * * : * *.:
Mb0715|M.bovis_AF2122/97 QLRDWIARKAFYGGSAAPLAVRHPDKTAPLVISGGALMAWILMSIGTGLG
Rv0696|M.tuberculosis_H37Rv QLRDWIARKAFYGGSAAPLAVRHPDKTAPLVISGGALMAWILMSIGTGLG
MMAR_1024|M.marinum_M QLRAWIARKAFYGGSAAPLSVRHPDKTAPLVISGWALTAWVLMSLGTRAT
MUL_0776|M.ulcerans_Agy99 QLRAWIARKAFYGGSAAPLSVRHPDKTAPLVISGWALTAWVLLSLGTRAT
MAV_4475|M.avium_104 ELRDWLARKAFYGGSAAPLSVRHPDKTAPVVISGWALMTWILMAFGSTLS
Mflv_5057|M.gilvum_PYR-GCK ELRKWFARKSFYGGSAAPLTIRHPGKTAPLVISGWMLVVWMLVAVGSGIG
Mvan_1299|M.vanbaalenii_PYR-1 ELRKWFARKSFYGSSAAPLTIRHPGKTAPLVISGWTLVVWMLVAIGSGIG
MSMEG_1426|M.smegmatis_MC2_155 NLRAWFHRKAFYGTSAAPLTVRHPGKTSPLVISGWTLMVWLMLGVGSFFG
TH_1903|M.thermoresistible__bu DIGEWLGRKAFYGSSAAPLTARHPDKTAPLLMSGWTLVLGVLVAMGSWVG
MAB_3831c|M.abscessus_ATCC_199 TLREWFARKAFYGKSAAPLSTRHPDKVAPMVISRWTLLVWILAAVGSGMG
MLBr_00752|M.leprae_Br4923 ----------------------HSTGRDPVSHLAAHHKSTYIFLADRHSG
*. *: : .
Mb0715|M.bovis_AF2122/97 RLASLVIAVLTGRRIARAMRCAETSFLDVLAVATRGLWAAALQLASAICR
Rv0696|M.tuberculosis_H37Rv RLASLVIAVLTGRRIARAMRCAETSFLDVLAVATRGLWAAALQLASAICR
MMAR_1024|M.marinum_M QLASLIIAVLTGRRIATALRSAQTSSWDVVVVAARGLWSAALQLASAICR
MUL_0776|M.ulcerans_Agy99 QLASLIIAVLTGRRIATALRSAQTSSWDVVVVAARGLWSAALQLASAICR
MAV_4475|M.avium_104 RLASLLLAVLTGRRIARAMRSAETSMTDVVTVAGRGLWSAALQLASALCR
Mflv_5057|M.gilvum_PYR-GCK YAASVAVAAVTGRRIAKSLSTVQTEPMEVAVVAAHGLWSAALQLASALCR
Mvan_1299|M.vanbaalenii_PYR-1 YFASIAVAAITGRRIAKSLASVQTEPMEVAVVAAHGLGSAALQLASAICR
MSMEG_1426|M.smegmatis_MC2_155 YLASLAAAVFAGTRIARALSVVETEPKEVAVVAAHGLWSSALQLCSAICR
TH_1903|M.thermoresistible__bu YLAALVLAVITGRRVAKSLSTVGSEPVEVVALAAQGLGSAARQLAGAICR
MAB_3831c|M.abscessus_ATCC_199 YLAAVGMAALAAGRVARTLRGVDTPPRDVVRVAAQGVGGAALQIASALCR
MLBr_00752|M.leprae_Br4923 WWRAPLRWMLRGSLVVRSRLMVRSSRRKQAAKQARQLAEGRR--------
: . . :. : . : . : : .
Mb0715|M.bovis_AF2122/97 HYWPLALLAAILSRRCRRVVLIAAVVDGVVDWLRRREGADDDAEPIGPLT
Rv0696|M.tuberculosis_H37Rv HYWPLALLAAILSRRCRRVVLIAAVVDGVVDWLRRREGADDDAEPIGPLT
MMAR_1024|M.marinum_M HYWPVTLIAVIVSRHCRRVVLVAAVIDGVVDWLRRREDPDGDSEPIGLLT
MUL_0776|M.ulcerans_Agy99 HYWPVTLIAVIVSRHCRRVVLVAAVIDGVVDWLRRREDPDGDSEPIGLLT
MAV_4475|M.avium_104 HYWPLALLAATMSRHFRRVVLVAAVMDGVVDWLRRRDAVGDDVEPIGLPT
Mflv_5057|M.gilvum_PYR-GCK HYWPIALVASVLSKRCRRVVVVAAVLDGVFDWVTRNGNADEDTKRVGILT
Mvan_1299|M.vanbaalenii_PYR-1 HYWPIALIAALVSRRSRRVVVVAAVLDGVFDWVTRNGNADEDTKRVGLPT
MSMEG_1426|M.smegmatis_MC2_155 HYWPIAMIAAVLFRRARHAVLVAAVVDGVVDWVTRRGNADDDTKPVGLLT
TH_1903|M.thermoresistible__bu HYWPIALIAALLSPRFRQVVVVAAVVDGVVDWLSRREHAGEDTKPVGLFA
MAB_3831c|M.abscessus_ATCC_199 HYWPLALFAAALSRRSRQVLVVAAIVDGVVDWMKRNDSSASPDDRIGLIE
MLBr_00752|M.leprae_Br4923 --------------------------------------------------
Mb0715|M.bovis_AF2122/97 YLVLKRVDDLAYGAGLWYGVVRERNIGALKPQIRT
Rv0696|M.tuberculosis_H37Rv YLVLKRVDDLAYGAGLWYGVVRERNIGALKPQIRT
MMAR_1024|M.marinum_M YLLLKRIDDLAYGLGLWYGVMRERNAGALKPQIRR
MUL_0776|M.ulcerans_Agy99 YLLLKRIDDLAYGLGLWYGVMRERNAGALKPQIRR
MAV_4475|M.avium_104 YLVLKRVDDLAYGLGLWWGVLRERNARALRPQIRS
Mflv_5057|M.gilvum_PYR-GCK YILLKRLDDIAYGLGLWSGVVRERHAGALKPQIRA
Mvan_1299|M.vanbaalenii_PYR-1 YLLLKRLDDIAYGLGLWTGVVRERHAGALKPQIRT
MSMEG_1426|M.smegmatis_MC2_155 HIVLKRLDDIAYGTGLWTGVVRERHLGALKPQVRS
TH_1903|M.thermoresistible__bu YLVLKRLDDLAYGCGLWAGVLRERDLGALKPQIRI
MAB_3831c|M.abscessus_ATCC_199 YVLLKRLDDIAYGIGLWSGIVRERDLGALRPELRP
MLBr_00752|M.leprae_Br4923 -----------------------------------