For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGNTRSDGLEAKAEALLALHRPGDPGVLPTVWDAWSARLAVDAGFAALTVGSHPVADSIGKSDGENMAFE DLLTRVRQITAAVEVPVSVDIESGYGESPERLIEGLLDAGVVGFNLEDTVHKEGKRLRTATEHAELVGQL RAAADAAGVHVVINARTDLFLRNDGDDADRVERAIARMSEAAAAGADSLFPAGLRDPEGLARFTSAVSLP VSVTVPPEDTDKAALAALGVGRITFGPFLQWALGAQAKEILGLWA
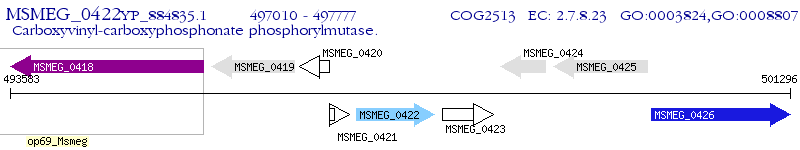
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0422 | - | - | 100% (255) | transferase |
| M. smegmatis MC2 155 | MSMEG_6646 | prpB | 1e-14 | 31.61% (193) | methylisocitrate lyase |
| M. smegmatis MC2 155 | MSMEG_4393 | - | 2e-12 | 30.92% (207) | carboxyvinyl-carboxyphosphonate phosphorylmutase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2021c | - | 4e-20 | 32.42% (219) | hypothetical protein Mb2021c |
| M. gilvum PYR-GCK | Mflv_0391 | - | 3e-94 | 67.20% (250) | hypothetical protein Mflv_0391 |
| M. tuberculosis H37Rv | Rv1998c | - | 4e-20 | 32.42% (219) | hypothetical protein Rv1998c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4410c | - | 1e-87 | 63.86% (249) | PEP phosphonomutase and related enzymes |
| M. marinum M | MMAR_0514 | - | 8e-92 | 66.80% (250) | PEP phosphonomutase |
| M. avium 104 | MAV_0345 | prpB | 2e-12 | 29.38% (211) | methylisocitrate lyase |
| M. thermoresistible (build 8) | TH_1918 | - | 6e-97 | 70.33% (246) | transferase |
| M. ulcerans Agy99 | MUL_1177 | - | 5e-83 | 65.79% (228) | PEP phosphonomutase |
| M. vanbaalenii PYR-1 | Mvan_0289 | - | 4e-97 | 69.08% (249) | hypothetical protein Mvan_0289 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2021c|M.bovis_AF2122/97 ---------------MSFHDLHHQGVPFVLPNAWDVPSALAYLAEGFTAI
Rv1998c|M.tuberculosis_H37Rv ---------------MSFHDLHHQGVPFVLPNAWDVPSALAYLAEGFTAI
Mflv_0391|M.gilvum_PYR-GCK ----MARTEDLKARAEALLALHQPGNPVILPTVWDAWSAHLAVDAGFSAL
Mvan_0289|M.vanbaalenii_PYR-1 -----MTNDVLKARAEALLALHQPGNPVILPTVWDAWSAKLATDAGFAAL
TH_1918|M.thermoresistible__bu ----------LQKRASDFLALHRPGDPVVLPTVWDAWSARLAVDAGFPAL
MMAR_0514|M.marinum_M -MPSEQGLNVTAERALALLALHQPGNPVVLPTVWDAWSARLAANAGFAAL
MUL_1177|M.ulcerans_Agy99 ----------------------------MLSTVWDAWSARLAANAGFAAL
MAB_4410c|M.abscessus_ATCC_199 ------MSTDLAAKASLLKSLHVPGDPVFLPTVWDAWSAKLAADAGFAAL
MSMEG_0422|M.smegmatis_MC2_155 --MGNTRSDGLEAKAEALLALHRPGDPGVLPTVWDAWSARLAVDAGFAAL
MAV_0345|M.avium_104 MRDMIGAARSPEQKRAQLRAALESGRLQRFPGAFSPLVAKLVAELGFDGV
:. .:. * ** .:
Mb2021c|M.bovis_AF2122/97 GTTSFGVSSSGGHPDGHRATRGANIA-LAAALAPLQCYVSVDIEDGYSD-
Rv1998c|M.tuberculosis_H37Rv GTTSFGVSSSGGHPDGHRATRGANIA-LAAALAPLQCYVSVDIEDGYSD-
Mflv_0391|M.gilvum_PYR-GCK TVGSHPLADSIGKPDNEGMTFDDVLTRVAQITAAVDVPLSVDIESGYAE-
Mvan_0289|M.vanbaalenii_PYR-1 TVGSHPLADSVGKSDREGMTFDDVLTRVAQITAAVDVPLSVDIESGYAE-
TH_1918|M.thermoresistible__bu TVGSHPVADSLGKADNEGMTFDDVLTRVGQITAAVEVPVSVDIESGYAE-
MMAR_0514|M.marinum_M TVGSHPLADSIGKPDNEGMSFDDVLARVTQITAAVDIPVSVDIESGYGL-
MUL_1177|M.ulcerans_Agy99 TVGSHPLADSIGKPDNEGMSFDDVLARVAQITAAVDIPVSVDIESGYGL-
MAB_4410c|M.abscessus_ATCC_199 TVGSHPLADSVGKADAEGMSFDDVVARVSQITAAVDLPVSVDIESGYAQ-
MSMEG_0422|M.smegmatis_MC2_155 TVGSHPVADSIGKSDGENMAFEDLLTRVRQITAAVEVPVSVDIESGYGE-
MAV_0345|M.avium_104 YVSGAALSADLGLPDIGLTTLTEVSGRGAQIAAVTELPTLIDADTGFGEP
. . :: . * .* : * : :* : *:.
Mb2021c|M.bovis_AF2122/97 -EPDAIADYVAQLSTAGINIED---SSAEKLIDP----ALAAAKIVAIKQ
Rv1998c|M.tuberculosis_H37Rv -EPDAIADYVAQLSTAGINIED---SSAEKLIDP----ALAAAKIVAIKQ
Mflv_0391|M.gilvum_PYR-GCK -EPGRLIDGLLSVDAVGLNIEDTVHSEGGRLRSSGEHAALVGGLRAAADA
Mvan_0289|M.vanbaalenii_PYR-1 -APTRLIEGLLSVGAVGLNIEDTVHSEGGRLRSSSEHAQLVGALRAAADA
TH_1918|M.thermoresistible__bu -SPERLITGLLDAGAVGLNIEDTVHSEGRRLRSVAEHAELVGALRAAADA
MMAR_0514|M.marinum_M -DAQRLIEGLLSVGAVGLNIEDTVHSEGARLRSRREHAELVGALRSAADA
MUL_1177|M.ulcerans_Agy99 -DAQRLIEGLLNVGAVGLNIEDTVHSEGARLRSRREHAELVGALRSAADA
MAB_4410c|M.abscessus_ATCC_199 -TAQRLIEGLIEAGAVGLNIEDTVHSEGGRLREPQEHADLVGALRVAADQ
MSMEG_0422|M.smegmatis_MC2_155 -SPERLIEGLLDAGVVGFNLEDTVHKEGKRLRTATEHAELVGQLRAAADA
MAV_0345|M.avium_104 LNAARTVTMLEYAGLAGCHLEDQVNPKRCGHLDG------KAVVPVDEMV
. : . .* ::** . .
Mb2021c|M.bovis_AF2122/97 RNPEVFVNARVDT---YWLRQHADTTSTIQRALR----YVDAGADGVFVP
Rv1998c|M.tuberculosis_H37Rv RNPEVFVNARVDT---YWLRQHADTTSTIQRALR----YVDAGADGVFVP
Mflv_0391|M.gilvum_PYR-GCK AGVHVVVNARTD----LFLRQDGDEADRVDRAIARLKEAADAGADVLYPV
Mvan_0289|M.vanbaalenii_PYR-1 AGVHVVVNARTD----LFLRQDGDESDRVDRAIARLKEAAAAGADVLYPV
TH_1918|M.thermoresistible__bu AGVHVVVNARTD----LFLRQDGDESDRVDRAIARLTECARAGADVLYPV
MMAR_0514|M.marinum_M AGVHVVLNARTD----LFLRRDGDPADRFERAVARLTEAAAAGADVLYPV
MUL_1177|M.ulcerans_Agy99 AGVHVVLNARTD----LFLRRDGDPADRFERAVAWLTEAAAAGADVLYPV
MAB_4410c|M.abscessus_ATCC_199 SGVPVVINARTD----VLLRQIGEESDRVDRAIARLKLAAEAGADSLYPV
MSMEG_0422|M.smegmatis_MC2_155 AGVHVVINARTD----LFLRNDGDDADRVERAIARMSEAAAAGADSLFPA
MAV_0345|M.avium_104 RRLRAAVSARRDPNFVVCARTDAAAIEGLPAAIERARAYADAGADLIFTE
. :.** * * . . . *: . **** ::
Mb2021c|M.bovis_AF2122/97 LANDPDELAELTRNIPCPVNT----LPVPG-LTIADLGELGVARVSTGSV
Rv1998c|M.tuberculosis_H37Rv LANDPDELAELTRNIPCPVNT----LPVPG-LTIADLGELGVARVSTGSV
Mflv_0391|M.gilvum_PYR-GCK GVHPPEVMRRLTSELPLPVNA----IARPDQSDPASYGPLGVARISFG--
Mvan_0289|M.vanbaalenii_PYR-1 GVHPPEVMRRLTSELPLPVNA----IARPESADPASYGPLGVARISFG--
TH_1918|M.thermoresistible__bu GRHDPDILRRLATELPLPVNA----IAAPDQADPASFGPLGVARISFG--
MMAR_0514|M.marinum_M GRHDPDTLRRLAAQLPLPINA----IALPDQDDPASFGRLGVARISFG--
MUL_1177|M.ulcerans_Agy99 GRHDPDTLRRLAAQLPLPINA----IALPDQDDPASFGRLGVARISVG--
MAB_4410c|M.abscessus_ATCC_199 GRHDPDVQRRLTSELPLPVNA----IAVPDVDDPASFGPLGVGRISFG--
MSMEG_0422|M.smegmatis_MC2_155 GLRDPEGLARFTSAVSLPVSV----TVPPEDTDKAALAALGVGRITFG--
MAV_0345|M.avium_104 ALQCPTEFERFRAALDTPLLANMTEFGKSPLLGADRLADIGYNVVIYPVT
. * .: : *: . . . :* :
Mb2021c|M.bovis_AF2122/97 PYSAGLYAAAHAARAVRDGEQLPRSVPYAELQARLVDYENRTSTT-----
Rv1998c|M.tuberculosis_H37Rv PYSAGLYAAAHAARAVSDGEQLPRSVPYAELQARLVDYENRTSTT-----
Mflv_0391|M.gilvum_PYR-GCK ----PFWQAALAERSK-------------ELLARWR--------------
Mvan_0289|M.vanbaalenii_PYR-1 ----PFWQAALAERAR-------------EILARWI--------------
TH_1918|M.thermoresistible__bu ----PFFQAALAVRAG-------------EILARWR--------------
MMAR_0514|M.marinum_M ----PFLQAALSGRAT-------------ELLARWR--------------
MUL_1177|M.ulcerans_Agy99 ----PFLQAGLSGRAT-------------ELLARWR--------------
MAB_4410c|M.abscessus_ATCC_199 ----PFFQRALSARAE-------------EILGRWR--------------
MSMEG_0422|M.smegmatis_MC2_155 ----PFLQWALGAQAK-------------EILGLWA--------------
MAV_0345|M.avium_104 TLRLAMHAVEAGLREIADTGTQSGLLDRMQHRSRLYQLLRYADYNHFDSD
: . : : .
Mb2021c|M.bovis_AF2122/97 ------
Rv1998c|M.tuberculosis_H37Rv ------
Mflv_0391|M.gilvum_PYR-GCK ------
Mvan_0289|M.vanbaalenii_PYR-1 ------
TH_1918|M.thermoresistible__bu ------
MMAR_0514|M.marinum_M ------
MUL_1177|M.ulcerans_Agy99 ------
MAB_4410c|M.abscessus_ATCC_199 ------
MSMEG_0422|M.smegmatis_MC2_155 ------
MAV_0345|M.avium_104 IYNFPR