For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRRLWIPLLVIAVVAVGGFTVSRLHNVFGAEKRPSYADTKAADSKPFDPKRLTYEVFGPPGTLADISYFD EDSNPQFVEKVSLPWSLHFDIGKTTAVGSIMAQGDSDTIGCRIIVDDEVKAEKVSNQTNAFTSCLLKAA
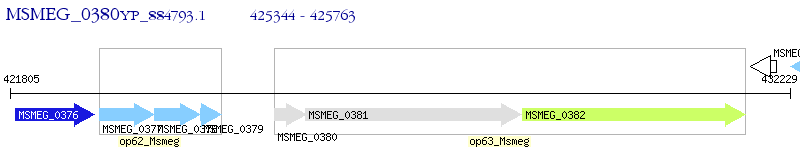
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0380 | - | - | 100% (139) | MmpS4 protein |
| M. smegmatis MC2 155 | MSMEG_3495 | - | 3e-37 | 52.90% (138) | MmpS5 protein |
| M. smegmatis MC2 155 | MSMEG_0226 | - | 2e-36 | 50.36% (139) | MmpS5 protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0459c | mmpS4 | 4e-48 | 60.58% (137) | membrane protein |
| M. gilvum PYR-GCK | Mflv_2704 | - | 1e-30 | 42.65% (136) | membrane family protein |
| M. tuberculosis H37Rv | Rv0451c | mmpS4 | 4e-48 | 60.58% (137) | membrane protein |
| M. leprae Br4923 | MLBr_02377 | - | 9e-46 | 56.93% (137) | hypothetical protein MLBr_02377 |
| M. abscessus ATCC 19977 | MAB_4117c | - | 5e-61 | 78.10% (137) | MmpS family protein |
| M. marinum M | MMAR_0772 | mmpS4_2 | 4e-44 | 56.93% (137) | membrane protein MmpS4_2 |
| M. avium 104 | MAV_3247 | - | 2e-46 | 58.57% (140) | MmpS4 protein |
| M. thermoresistible (build 8) | TH_3673 | - | 4e-54 | 68.35% (139) | PUTATIVE MmpS4 protein |
| M. ulcerans Agy99 | MUL_2383 | mmpS4 | 4e-31 | 42.86% (140) | membrane protein MmpS4 |
| M. vanbaalenii PYR-1 | Mvan_3834 | - | 1e-33 | 43.17% (139) | hypothetical protein Mvan_3834 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0380|M.smegmatis_MC2_155 ----------------MRRLWIPLLVIAVVAVGGFTVSRLHNVFGAEKRP
MAB_4117c|M.abscessus_ATCC_199 ----------------MR-LWIPLLVIAVIAVGGFTVSRLHGVFGNEKRP
TH_3673|M.thermoresistible__bu ----------------LKRFWIPLLALVVFSSGGFAVSRLHGIFGSEKRP
Mb0459c|M.bovis_AF2122/97 ---------------MLMRTWIPLVILVVVIVGGFTVHRIRGFFGSENRP
Rv0451c|M.tuberculosis_H37Rv ---------------MLMRTWIPLVILVVVIVGGFTVHRIRGFFGSENRP
MMAR_0772|M.marinum_M ---------------MLSRTWIVLVIVAVIIVGGFSVHRIRGFFASEKRE
MLBr_02377|M.leprae_Br4923 -MSKRQRGKGISIFKLLSRIWIPLVILVVLVVGGFVVYRVHSYFASEKRE
MAV_3247|M.avium_104 ----------------MKRLWIPLVIVAVVVAGGLTVSRLHGIFGSEKRP
Mflv_2704|M.gilvum_PYR-GCK ----------------MSRAWIPLVLVIVLLVAGFTVWRLRNMFASHPLP
Mvan_3834|M.vanbaalenii_PYR-1 ----------------MTRAWIPLVLVLVLVVGGLAVWRIRNIFGSHQLP
MUL_2383|M.ulcerans_Agy99 MPAGPRGGGCTEIVTVLKRAWLPLVLAVVFAVGGFAVSRVRGMFGSQQLP
: *: *: *. .*: * *::. *. .
MSMEG_0380|M.smegmatis_MC2_155 SYA-DTKAADSKPFDPKRLTYEVFGPPGTLADISYFDEDSNPQFVEKVSL
MAB_4117c|M.abscessus_ATCC_199 SYA-DTNVSDAKSFDPKKMSYEVFGSPGNVADISYFDVNGDPLFIQKVPL
TH_3673|M.thermoresistible__bu TYA-AALQNEVTTFNPKRLTYEVFGPPGTTASISYFDENTDPQFIENVAL
Mb0459c|M.bovis_AF2122/97 SYS-DTNLENSKPFNPKHLTYEIFGPPGTVADISYFDVNSEPQRVDGAVL
Rv0451c|M.tuberculosis_H37Rv SYS-DTNLENSKPFNPKHLTYEIFGPPGTVADISYFDVNSEPQRVDGAVL
MMAR_0772|M.marinum_M SYS-DSNLDN-KPFNPKEITYEVFGPPGTVADISYFDVNSDPQQVEGAVL
MLBr_02377|M.leprae_Br4923 SYA-DSNLGSSKPFNPKQIVYEVFGPPGTVADISYFDANSDPQRIDGAQL
MAV_3247|M.avium_104 SYA-DTRQQDTKPFNPKHVKYEVFGPAGAMADISYFDANGEPQHINGVEL
Mflv_2704|M.gilvum_PYR-GCK TYAGSMSA-DTNKSDPKIVRYEIFGVPGATADINYIDDEGDANQIIGAPL
Mvan_3834|M.vanbaalenii_PYR-1 TYAGSMSE-DSNKSDPKIVRYEIFGDPGATADINYIDDEGDPNQIVGATL
MUL_2383|M.ulcerans_Agy99 TYADSTPAGSVSSSGPKRVRYEIFGAPGTIADINYMDPAGEAQQLNDVAL
:*: . . .** : **:** .* *.*.*:* :. : . *
MSMEG_0380|M.smegmatis_MC2_155 PWSLHFDIGKTTAVGSIMAQGDSDTIGCRIIVDDEVKAEKVSNQTNAFTS
MAB_4117c|M.abscessus_ATCC_199 PWSVKFEIGKTTAVGSIMAQGDGSSIGCRIIVDDEVKSEKVTNQTNAFTS
TH_3673|M.thermoresistible__bu PWSLQMEISKTTALGSIMAQGDSNSIGCRIMVDDEVKEEKVSNQVNAFAS
Mb0459c|M.bovis_AF2122/97 PWSLHITTNDAAVMGNIVAQGNSDSIGCRITVDGKVRAERVSNEVNAYTY
Rv0451c|M.tuberculosis_H37Rv PWSLHITTNDAAVMGNIVAQGNSDSIGCRITVDGKVRAERVSNEVNAYTY
MMAR_0772|M.marinum_M PWTLHFTTNLAAVMGNLVAQGNTNSIGCRITVDGKVKAERVSNEVNAYTY
MLBr_02377|M.leprae_Br4923 PWSLLMTTTLAAVMGNLVAQGNTDSIGCRIIVDGVVKAERVSNEVNAYTY
MAV_3247|M.avium_104 PWTFDISTTLPSIVGNVVAQGNSDSLGCRILVDGVVKTERISHELNAFTY
Mflv_2704|M.gilvum_PYR-GCK PWAIEVETDSPAMVGNIVAQGDGDFIGCRILSDGELKDERTIHNYTAYIY
Mvan_3834|M.vanbaalenii_PYR-1 PWSIEVETNSPAMVGNVVAQGDSDFIGCRISADGVVKDERTAHNVSAYVY
MUL_2383|M.ulcerans_Agy99 PWSIEVETDAPSMVANIIAQANTDFLGCRISSGGEVKDERTSSAVSAYVY
**:. . .: :..::**.: . :**** .. :: *: .*:
MSMEG_0380|M.smegmatis_MC2_155 CLLKAA
MAB_4117c|M.abscessus_ATCC_199 CLLKAA
TH_3673|M.thermoresistible__bu CLLQAA
Mb0459c|M.bovis_AF2122/97 CLVKSA
Rv0451c|M.tuberculosis_H37Rv CLVKSA
MMAR_0772|M.marinum_M CLVKSA
MLBr_02377|M.leprae_Br4923 CLVKSA
MAV_3247|M.avium_104 CVLTAT
Mflv_2704|M.gilvum_PYR-GCK CFSKAT
Mvan_3834|M.vanbaalenii_PYR-1 CFTKSA
MUL_2383|M.ulcerans_Agy99 CIAKSA
*. ::