For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SALTAPNVTVDKTLIHRFTEAVAGHAEVITDGAVLDARGHDFWGVGGTAELLLHPHDRDEVAAIMRIASQ HQVPIVPRGGASNCSGGMMPARGRVLLDLSNLNRIIDVDVESRCARVEPGVINSALQEHLAPYGLCFSPD PVSAHLATVGGNIIENAGGPHALKYGVTYNHILGVEVVLPDGTTVTLDAEDDGPDLLGLVVGSEGTLGIV TEATVALRPVAEVTHSLMGAFATAREAADTIAAIIATGVVPAAVEWLDHDGIMGLQQFYDTGYPLDADSI VLIDVDGTADEVARDQAIVERVLRERATTVRIAEDEKDRAALWYGRLHAPDSVVQSGKGFFIGDVTVPRD RIPEMQEAIGATAERHKDGLLFIAVCGHAGDGDLHPTTFYDKDNPLAASALEAANNEIIEAALRLGGTIT GEHGVGTEKIQFMTKRFTPVEIAAQRAIKEAFDPAGLLNPGVMLPEQSPDEPVTAAFGTAVRDAVEGTLL TDPGAALTTGTDTGIAANLGNLSLVVGADATVEQINRYLDDHGIFCPAVPATGGERSIGEVVATATGAER IGIRHALLGADVTVVDGNAAARFGAETMKDVAGYDAKRLYIGGHGAFGALRTLIFKICVRR*
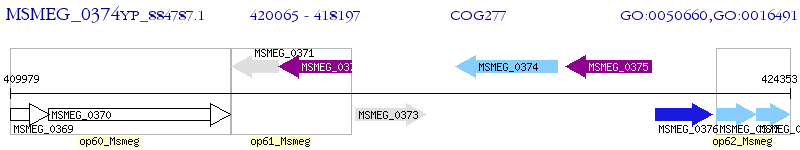
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0374 | - | - | 100% (622) | glycolate oxidase subunit |
| M. smegmatis MC2 155 | MSMEG_0301 | - | 2e-66 | 35.45% (409) | oxidoreductase, FAD-binding |
| M. smegmatis MC2 155 | MSMEG_5037 | - | 1e-61 | 33.73% (418) | oxidoreductase, FAD-binding |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2301 | - | 1e-53 | 31.00% (471) | dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2230 | - | 4e-64 | 33.18% (443) | FAD linked oxidase domain-containing protein |
| M. tuberculosis H37Rv | Rv1257c | - | 7e-64 | 34.09% (440) | oxidoreductase |
| M. leprae Br4923 | MLBr_01103 | - | 6e-62 | 32.60% (454) | putative oxidoreductase subunit |
| M. abscessus ATCC 19977 | MAB_1407c | - | 7e-67 | 33.33% (462) | oxidoreductase |
| M. marinum M | MMAR_4183 | - | 2e-57 | 31.82% (440) | oxidoreductase |
| M. avium 104 | MAV_1405 | - | 9e-60 | 33.18% (440) | oxidoreductase, FAD-binding |
| M. thermoresistible (build 8) | TH_3358 | - | 3e-65 | 34.61% (419) | PUTATIVE FAD linked oxidase domain protein |
| M. ulcerans Agy99 | MUL_4485 | - | 2e-57 | 31.59% (459) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_4464 | - | 6e-63 | 33.81% (423) | FAD linked oxidase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4183|M.marinum_M ----------MNADVLASLIAGLP-DGMVVTDPTVTEGYRQDRALDPSAG
MUL_4485|M.ulcerans_Agy99 ----------MNADVLASLIAGLP-DGMAVTDPTVTEGYRQDRALDPSAG
MAV_1405|M.avium_104 ----------MSSEALASLIADLP-DGTVVTDPTVTEGYRQDRAFDPSAG
Rv1257c|M.tuberculosis_H37Rv ----------MNTDVLAGLMAELP-EGMVVTDPAVTDGYRQDRAFDPSAG
MLBr_01103|M.leprae_Br4923 ------------MGVLTSLIAGLP-DGMVVTDSAVTDGYRHDRALDPSAG
Mflv_2230|M.gilvum_PYR-GCK ------------MQPLASLIADLP-DGTVVTDPDILESYRQDRAADPAAG
Mvan_4464|M.vanbaalenii_PYR-1 ------------MQPLASLIAELP-DGTVVTDPDIVASYRQDRAADPSAG
TH_3358|M.thermoresistible__bu -----------VSAALSALIAELP-DGTVVTDPDILASYRQDRARDPDAG
MAB_1407c|M.abscessus_ATCC_199 ----------MSRDALHALTAVLG-EGAVITDPDITASYRQDRAFDPDAG
Mb2301|M.bovis_AF2122/97 -----------MSEMTARFSEIVG-NANLLTGDAIPEDYAHDEELTGPPQ
MSMEG_0374|M.smegmatis_MC2_155 MSALTAPNVTVDKTLIHRFTEAVAGHAEVITDGAVLDARGHD--FWGVGG
: : .. :*. : :*
MMAR_4183|M.marinum_M KPLAVVRPRCTEEVQTILRWAGANRVAVVTRGAGSGLSGGATALDGGIVL
MUL_4485|M.ulcerans_Agy99 KPLAVVRPRCTEEVQTILRWAGANRVAVVTRGAGSGLSGGATALDGGIVL
MAV_1405|M.avium_104 KPLAVVRPRRTEEVQTVLRWASAHGVPVVTRGAGSGLSGGATALDNGIVL
Rv1257c|M.tuberculosis_H37Rv KPLAIIRPRRTEEVQTVLRWASANQVPVVTRGAGSGLSGGATALDGGIVL
MLBr_01103|M.leprae_Br4923 KPLAVVQPRRTDEVQTVLRWASANRVPVVARGAGSGLSGGATALDDGIVL
Mflv_2230|M.gilvum_PYR-GCK TPLAVVRPRRTEEVQTVLRWAGTHRIAVVPRGMGTGLSGGATALDGGIVL
Mvan_4464|M.vanbaalenii_PYR-1 TPIAVVRPRRTEEVQATLRWATAHRIAVVPRGMGTGLSGGATALDGGIVL
TH_3358|M.thermoresistible__bu TPLAVVRPRRTEEVQTVLRWANAHRVPVVPRGAGTGLSGGASALDGGIVL
MAB_1407c|M.abscessus_ATCC_199 MPLAVVRPRSTEEVSATLKWASAHHISVVPRGAGTGLSGGSTAQDGAVVL
Mb2301|M.bovis_AF2122/97 KPAYAAKPATPEEVAQLLKAASENGVPVTARGSGCGLSGAARPVEGGLLI
MSMEG_0374|M.smegmatis_MC2_155 TAELLLHPHDRDEVAAIMRIASQHQVPIVPRGGASNCSGGMMPARGRVLL
. :* :** :: * : :.:..** . . **. . . :::
MMAR_4183|M.marinum_M STEKMR-DITIDPVTRIAVCQPGLFNAEVKKAAAEHGLWYPPDPSSFEIC
MUL_4485|M.ulcerans_Agy99 STEKMR-DITIDPVTRTAVCQPGLFNAEVKKAAAEHGLWYPPDPSSFEIC
MAV_1405|M.avium_104 STEKMR-DITVDPVTRTAVCQPGLFNAEVKKAAAEHGLWYPPDPSSFEIC
Rv1257c|M.tuberculosis_H37Rv STEKMR-DITVDPVTRTAVCQPGLYNAEVKEAAAEHGLWYPPDPSSFEIC
MLBr_01103|M.leprae_Br4923 STEKMR-DITVDPVTRTAVCQPGLFNAEVKKAAAEHGLWYPPDPSSFEIC
Mflv_2230|M.gilvum_PYR-GCK STEKMR-DITVDPVTRTAVVQPGLLNAEVKKVVAEHGLWYPPDPSSFEIC
Mvan_4464|M.vanbaalenii_PYR-1 STEKMR-DITVDPVTRTAVAQPGLLNAEVKKAVAEYGLWYPPDPSSFEIC
TH_3358|M.thermoresistible__bu STEKMR-DITVDPVTRTAVVQPGLLNAEVKQAAAEHGLWYPPDPSSFEIC
MAB_1407c|M.abscessus_ATCC_199 STERMR-TITIDTATRVAITQPGALNAEVKAAAAAQGLWYPPDPSSFEIC
Mb2301|M.bovis_AF2122/97 SFDRMNKVLEVDTANQVAVVQPGVALTDLDAATADTGLRYTVYPGELSSS
MSMEG_0374|M.smegmatis_MC2_155 DLSNLNRIIDVDVESRCARVEPGVINSALQEHLAPYGLCFSPDPVSAHLA
. ..:. : :* .: * :** : :. * ** :. * . .
MMAR_4183|M.marinum_M SIGGNIATNAGGLCCVKYGVTTDYVLGMQVVLADGTAVRLGGPRLKDVAG
MUL_4485|M.ulcerans_Agy99 SIGGNIATNAGGLCCVKYGVTTDYVLGMQVVLADGTAVRLGGPRLKDVAG
MAV_1405|M.avium_104 SIGGNIATNAGGLCCVKYGVTTDYVLGMQVVLADGTAVRLGGPRLKDVAG
Rv1257c|M.tuberculosis_H37Rv SIGGNIATNAGGLCCVKYGVTGDYVLGMQVVLANGTAVRLGGPRLKDVAG
MLBr_01103|M.leprae_Br4923 SIGGNVATNAGGLCCVKYGVTGDYVLGIQVVLADGTAVRLGGPRLKDVAG
Mflv_2230|M.gilvum_PYR-GCK SIGGNIATNAGGLCCVKYGVTTDYVLGLQVVLADGTAVRLGGPRLKDVAG
Mvan_4464|M.vanbaalenii_PYR-1 SIGGNIATNAGGLCCVKYGVTTDYVLGLQVVLADGTAVRLGGPRLKDVAG
TH_3358|M.thermoresistible__bu SIGGNIATNAGGLCCVKYGVTTDYVMGLQVVLADGTAVRLGGPRIKDVAG
MAB_1407c|M.abscessus_ATCC_199 SIGGNIATNAGGLCCVKYGVTTDYVLGMQVVLADGTVLKLGGPRLKDVAG
Mb2301|M.bovis_AF2122/97 -VGGNVGTNAGGMRAVKYGVARHNVLGLQAVLPTGEIIRTGGRMAKVSTG
MSMEG_0374|M.smegmatis_MC2_155 TVGGNIIENAGGPHALKYGVTYNHILGVEVVLPDGTTVTLDAE----DDG
:***: **** .:****: . ::*::.**. * : .. *
MMAR_4183|M.marinum_M LSLTKLFVGSEGTLGVITEVTLRLLPAQNAAAVVVASFASVESAVDAVLG
MUL_4485|M.ulcerans_Agy99 LSLTKLFVGSEGTLGVITEVTLRLLPAQNAAAVVVASFASVESAVDAVLG
MAV_1405|M.avium_104 LSLTKLFVGSEGTLGVITEVTLRLVPKQNTSSVVVASFATVEDAVGAVLG
Rv1257c|M.tuberculosis_H37Rv LSLTKLFVGSEGTLGVITEVTLRLLPAQNASSIVVASFGSVQAAVDAVLG
MLBr_01103|M.leprae_Br4923 LSLTKLFVGSEGTLGIITEVTLRLLPTQNTSSIVVATFASVQAAVDAVLG
Mflv_2230|M.gilvum_PYR-GCK LSLTKLFVGSEGTLGVVTEVTLKLLPAQHPACTVVATFDSVESAADAVVE
Mvan_4464|M.vanbaalenii_PYR-1 LSLTKLFVGSEGTLGVVTEVTLKLLPAQSGACTVVATFDSVEDAANAVVT
TH_3358|M.thermoresistible__bu LPLTKLFVGSEGTLGVVTEVTLRLLPPQHQASTVVATFASVEAAAESVLA
MAB_1407c|M.abscessus_ATCC_199 LSLTKLFVGSEGTLGVITEVTLRLLPCQPTPCTVVATFSSVHDATDAVLA
Mb2301|M.bovis_AF2122/97 YDLTQLIIGSEGTLALVTEVIVKLHPRLDHNASVLAPFADFDQVMAAVPK
MSMEG_0374|M.smegmatis_MC2_155 PDLLGLVVGSEGTLGIVTEATVALRPVAEVTHSLMGAFATAREAADTIAA
* *.:******.::**. : * * ::..* . ::
MMAR_4183|M.marinum_M VSAR-IRPSMLEFMDSVAINAVEDTLRMDL-------DRGAAAMLVASSD
MUL_4485|M.ulcerans_Agy99 VSAR-IRPSMLEFMDSVAINAVEDTLRMDL-------DRGAAAMLVASSD
MAV_1405|M.avium_104 VTGR-LRPSMLEFMDSVAINAVEDTLRMDL-------DRGAAAMLVAGSD
Rv1257c|M.tuberculosis_H37Rv VTGR-LRPAMLEFMDSVAINAVEDTLRMDL-------DRDAAAMLVAGSD
MLBr_01103|M.leprae_Br4923 VAAR-LRPAMLEFMDSVAINAVEDTFRMGL-------DREAAAMLVAGSD
Mflv_2230|M.gilvum_PYR-GCK ITGR-IRPSMLEFMDSTAINAVEDKLKMGL-------DRDAAAMMVAASD
Mvan_4464|M.vanbaalenii_PYR-1 ITGK-IRPSMLEFMDSAAINAVEDKLKMGL-------DRSAAAMMVAASD
TH_3358|M.thermoresistible__bu ITAR-IRPSMLEFMDAVAINAVEDKLRMGL-------DRSAAAMMVAASD
MAB_1407c|M.abscessus_ATCC_199 ITAQ-IRPSMLEFMDSVAINAVEDKLKMGL-------DRAAGAMLVARSD
Mb2301|M.bovis_AF2122/97 ILASGLAPDILEYIDNTSMAALISTQNLELGIPDQIRDSCEAYLLVALEN
MSMEG_0374|M.smegmatis_MC2_155 IIATGVVPAAVEWLDHDGIMGLQQFYDTGY-------PLDADSIVLIDVD
: . : * :*::* .: .: . ::: :
MMAR_4183|M.marinum_M ERGQAGRDDAELMAAVFAEHGATEVFSTTDPDEGEAFVAARRFCIPAVEA
MUL_4485|M.ulcerans_Agy99 ERGQADRDDAELMAAVFAEHGATEVFSTTDPDEGEPFVAARRFCIPAVEA
MAV_1405|M.avium_104 ERGRAGSEDAEIMAKVFAENAATEVFSTDDPDEGEAFVVARRFCIPAVEA
Rv1257c|M.tuberculosis_H37Rv ERGRAATEDAAVMAAVFAENGAIDVFSTDDPDEGEAFIAARRFAIPAVES
MLBr_01103|M.leprae_Br4923 ELGHAGTDEAKLIGAVFAENGATQVSSTNDPDEGEAFVAARRFCIPAVES
Mflv_2230|M.gilvum_PYR-GCK DRGAAGAQDAEYMARVFTEHGATEVFSTSDPDEGDAFVAARRFAIPAVES
Mvan_4464|M.vanbaalenii_PYR-1 DRGPSGAQDAEFMAGVFTEHGAREVFSTSDPDEGEAFVAARRFAIPAVEA
TH_3358|M.thermoresistible__bu DRGPAGAEDAEFMAQVFTEHGATEVFSTADPDEGEAFVAARRFAIPAVEA
MAB_1407c|M.abscessus_ATCC_199 ERGHTSTEDAELIARIFTDHHAAEVFSTDDEEEGEAFVAARRFAIPAVEA
Mb2301|M.bovis_AF2122/97 RIADRLFEDIQTVGEMLMELGAVDAYVLEG-GSARKLIEAREKAFWAAKA
MSMEG_0374|M.smegmatis_MC2_155 GTADEVARDQAIVERVLRERATTVRIAEDEKDRAALWYGRLHAPDSVVQS
. : : :: : : . . ..::
MMAR_4183|M.marinum_M KGSLLLEDVGVPLPNLGKLVTGIARIAQERD---LMISVIAHAGDGNTHP
MUL_4485|M.ulcerans_Agy99 KGSLLLEDVGVPLPNLRKLVTGIARIAQERD---LMISVIAHAGDGNTHP
MAV_1405|M.avium_104 KGSLLLEDVGVPLPALGRLVTGIAAIAAERD---LMISVIAHAGDGNTHP
Rv1257c|M.tuberculosis_H37Rv KGALLLEDVGVPLPALGELVTGIARIAEERN---LMISVIAHAGDGNTHP
MLBr_01103|M.leprae_Br4923 KGSLLLEDVGVPLPALGKLVTGIAHISTERN---LMISVIAHAGDGNTHP
Mflv_2230|M.gilvum_PYR-GCK KGSLLLEDVGVPLPALAELVGGVEKIAAARD---VMISVIAHAGDGNTHP
Mvan_4464|M.vanbaalenii_PYR-1 RGALLLEDVGVPLPALAELVGGVEKIAGHHE---LMISVIAHAGDGNTHP
TH_3358|M.thermoresistible__bu KGALLLEDVGVPLPALADLVGGVARLADHHE---LTISVIAHAGDGNTHP
MAB_1407c|M.abscessus_ATCC_199 KGSLLLEDVGVPLPKLAQLVEGVEAIAAQHR---LLISVIAHAGDGNTHP
Mb2301|M.bovis_AF2122/97 LGADDIIDTVVPRASMPKFLSTARGLAAAAD---GAAVGCGHAGDGNVHM
MSMEG_0374|M.smegmatis_MC2_155 GKGFFIGDVTVPRDRIPEMQEAIGATAERHKDGLLFIAVCGHAGDGDLHP
. : *. ** : : : .*****: *
MMAR_4183|M.marinum_M LLVYDPSDAAMAERAQLAYGEIMDLAVSLGGTITGEHGVGRLKRPWLEGY
MUL_4485|M.ulcerans_Agy99 LLVYDPSDAAMAERAQLAYGEIMDLAVGLGGTITGEHGVGRLKRPWLEGY
MAV_1405|M.avium_104 LIVYDPADADMTERAHLAFGEIMDLAVGLGGTITGEHGVGRLKRPWLAGY
Rv1257c|M.tuberculosis_H37Rv LLVYDPADAAMLERAHLAYGEIMDLAVGLGGTITGEHGVGRLKRPWLAGY
MLBr_01103|M.leprae_Br4923 LIVYNPADATMTERAHRAFGEIMDMAVGLGGTITGEHGVGRLKRPWLDDY
Mflv_2230|M.gilvum_PYR-GCK LIVFDPSDTAMEQRAQQAFGEIMDLAVSLGGTITGEHGVGRLKRPWLAGQ
Mvan_4464|M.vanbaalenii_PYR-1 LIVFDPDDPDMERRAQQAFGEIMDLAIGLGGTITGEHGVGRLKRPWLAGQ
TH_3358|M.thermoresistible__bu LIVYDPADAAMAERAAQAFGEIMELAIDLGGTITGEHGVGRLKRPWLADQ
MAB_1407c|M.abscessus_ATCC_199 LIVHDPTDPDESRRAQTAFGQIMDLAISLGGTITGEHGVGRLKKPWLGDQ
Mb2301|M.bovis_AF2122/97 AIACKDP-----EKKKKLMTDIFALAMELGGAISGEHGVGRAKTGYFLEL
MSMEG_0374|M.smegmatis_MC2_155 TTFYDKDNPLAASALEAANNEIIEAALRLGGTITGEHGVGTEKIQFMTKR
. :*: *: ***:*:****** * ::
MMAR_4183|M.marinum_M LGPDVMDLNRRIKQALDPLDILNPGSGI----------------------
MUL_4485|M.ulcerans_Agy99 LGPDVMDLNRRIKQALDPLDILNPGSGI----------------------
MAV_1405|M.avium_104 LGPEVMDLNRRIKQALDPQGILNPGSGI----------------------
Rv1257c|M.tuberculosis_H37Rv LGPDVLALNQRIKQALDPQGILNPGSAI----------------------
MLBr_01103|M.leprae_Br4923 LGPDVMELNQRIKHALDPYGILNPGAAI----------------------
Mflv_2230|M.gilvum_PYR-GCK LGPEAMELNRRIKQALDPDGILNPGAAI----------------------
Mvan_4464|M.vanbaalenii_PYR-1 LGPEAMELNRRIKQALDPDGILNPGAAI----------------------
TH_3358|M.thermoresistible__bu IGPEAMELHRRIKDALDPNGILNPGAGF----------------------
MAB_1407c|M.abscessus_ATCC_199 VGPEVLEVNRRIKQALDPDGILNPGTLI----------------------
Mb2301|M.bovis_AF2122/97 EDPVKISLMRRIKQSFDPAGILNPGVVFGDT-------------------
MSMEG_0374|M.smegmatis_MC2_155 FTPVEIAAQRAIKEAFDPAGLLNPGVMLPEQSPDEPVTAAFGTAVRDAVE
* : : **.::** .:**** :
MMAR_4183|M.marinum_M --------------------------------------------------
MUL_4485|M.ulcerans_Agy99 --------------------------------------------------
MAV_1405|M.avium_104 --------------------------------------------------
Rv1257c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01103|M.leprae_Br4923 --------------------------------------------------
Mflv_2230|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4464|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3358|M.thermoresistible__bu --------------------------------------------------
MAB_1407c|M.abscessus_ATCC_199 --------------------------------------------------
Mb2301|M.bovis_AF2122/97 --------------------------------------------------
MSMEG_0374|M.smegmatis_MC2_155 GTLLTDPGAALTTGTDTGIAANLGNLSLVVGADATVEQINRYLDDHGIFC
MMAR_4183|M.marinum_M --------------------------------------------------
MUL_4485|M.ulcerans_Agy99 --------------------------------------------------
MAV_1405|M.avium_104 --------------------------------------------------
Rv1257c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01103|M.leprae_Br4923 --------------------------------------------------
Mflv_2230|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4464|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3358|M.thermoresistible__bu --------------------------------------------------
MAB_1407c|M.abscessus_ATCC_199 --------------------------------------------------
Mb2301|M.bovis_AF2122/97 --------------------------------------------------
MSMEG_0374|M.smegmatis_MC2_155 PAVPATGGERSIGEVVATATGAERIGIRHALLGADVTVVDGNAAARFGAE
MMAR_4183|M.marinum_M -----------------------------------
MUL_4485|M.ulcerans_Agy99 -----------------------------------
MAV_1405|M.avium_104 -----------------------------------
Rv1257c|M.tuberculosis_H37Rv -----------------------------------
MLBr_01103|M.leprae_Br4923 -----------------------------------
Mflv_2230|M.gilvum_PYR-GCK -----------------------------------
Mvan_4464|M.vanbaalenii_PYR-1 -----------------------------------
TH_3358|M.thermoresistible__bu -----------------------------------
MAB_1407c|M.abscessus_ATCC_199 -----------------------------------
Mb2301|M.bovis_AF2122/97 -----------------------------------
MSMEG_0374|M.smegmatis_MC2_155 TMKDVAGYDAKRLYIGGHGAFGALRTLIFKICVRR