For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPPLTSEIDPHSDTFKSNYEVQTAAVAALNEQLAIVAGGGGERYVKRHHDRGRLLARERIELLLDRDAPF LELSSLAAWGTDFNVGAAIVTGVGVVSGTECMIIAHDPTVRGGTMNPYTLRKNLRALEIAKVNRLPVLYL VESGGADLPTQSELFVHAGRIFRDLTELSSMGIPSVAMVFGNSTAGGAYVPGMCDYAVLVDKQAKVFLGG PPLVKMATGEDADDESLGGADMHTRVSGLGDYFAVDEVDAIRIGRDIVAHLNWRKLGPGPSRPADEPLYD SEELLGIASGDSRVPFDPREVIARIVDGSRFSEYKPRWGTSLATGWASIHGFPVGILANTRGVLFSEESK KATEFILLANQTDTPLIFLQNVTGFMVGTEYEQGGIIKDGAKMINAVTNSTVPHITLNMGGSFGAGNYGM SGRAYDPRFMFSWVNAKLAVMGAQQLAGVLSIVGKAAAQAAGREFDHEADAKRTKEIEEQIDRESHAFFN SARIYDDGVIDPRDTRSVLGIALSAAHSNVVAGRRGFGVFRM
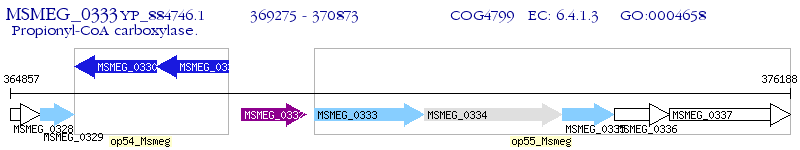
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0333 | - | - | 100% (532) | carboxyl transferase domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_5492 | - | 0.0 | 65.04% (532) | acetyl-CoA carboxylase carboxyltransferase |
| M. smegmatis MC2 155 | MSMEG_4717 | - | e-136 | 50.10% (521) | carboxyl transferase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0999c | accD2 | 0.0 | 63.52% (529) | acetyl-/propionyl-CoA carboxylase subunit beta |
| M. gilvum PYR-GCK | Mflv_4613 | - | 0.0 | 65.78% (529) | carboxyl transferase |
| M. tuberculosis H37Rv | Rv0974c | accD2 | 0.0 | 63.52% (529) | acetyl-/propionyl-CoA carboxylase subunit beta |
| M. leprae Br4923 | MLBr_00731 | accD5 | 1e-57 | 30.87% (515) | acetyl/propionyl CoA carboxylase beta subunit |
| M. abscessus ATCC 19977 | MAB_2067c | - | 0.0 | 66.35% (529) | acetyl/propionyl-CoA carboxylase beta subunit AccD2 |
| M. marinum M | MMAR_4533 | accD2 | 0.0 | 64.65% (529) | acetyl-/propionyl-CoA carboxylase (beta subunit) AccD2 |
| M. avium 104 | MAV_1090 | - | 0.0 | 64.47% (532) | acetyl-CoA carboxylase carboxyltransferase |
| M. thermoresistible (build 8) | TH_2815 | - | 1e-133 | 50.29% (509) | PUTATIVE Carboxyl transferase domain protein |
| M. ulcerans Agy99 | MUL_4705 | accD2 | 0.0 | 64.08% (529) | acetyl-/propionyl-CoA carboxylase (beta subunit) AccD2 |
| M. vanbaalenii PYR-1 | Mvan_0346 | - | 0.0 | 90.79% (532) | carboxyl transferase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0333|M.smegmatis_MC2_155 -------MPPLTSEIDPHSDTFKSNYEVQTAAVAALNEQLAIVAGGGGER
Mvan_0346|M.vanbaalenii_PYR-1 -------MTVLTSDVDPASDTFQFNREVQSAAVAALDEQLRLVADGGGER
Mb0999c|M.bovis_AF2122/97 ---------MLQSTLDPNASAYDEAAATMSGKLDEINAELAKALAGGGPK
Rv0974c|M.tuberculosis_H37Rv ---------MLQSTLDPNASAYDEAAATMSGKLDEINAELAKALAGGGPK
MMAR_4533|M.marinum_M -------MTVLRSTLDPNGAAFSEAASAATTKLDEIDAELANALAGGGPK
MUL_4705|M.ulcerans_Agy99 -----MRVTVLRSTLDPNGAAFSEAASAATTKLDEIDAELANALAGGGPK
MAV_1090|M.avium_104 -------MTVLQSTLDREATAFTDAASAAMAKLEEIDTELANALAGGGPK
Mflv_4613|M.gilvum_PYR-GCK MSPGGKLIDVLPDRVDTRAPVYLENREGLIAQLHALAEQLALANGGGGEK
MAB_2067c|M.abscessus_ATCC_199 ------MVQVLPDRVDDTAPGYLKNREGLSAQLDTIAEQLALVNGGGGAK
TH_2815|M.thermoresistible__bu -----------------------------------LRSKLAAAARGGPER
MLBr_00731|M.leprae_Br4923 ----MTSVTDHSAHSMERAAEHTINIHTTAGKLAELHKRTEEALHPVGAA
: . .
MSMEG_0333|M.smegmatis_MC2_155 YVKRHHDRGRLLARERIELLLDRDAPFLELSSLAAWGT-------DFNVG
Mvan_0346|M.vanbaalenii_PYR-1 YVKRHHDRGRLLARERIELLVDRDAPFMELSALAAWGT-------EFNVG
Mb0999c|M.bovis_AF2122/97 YVDRHHARGKLTPRERIELLVDPDSPFLELSPLAAYGS-------NFQIG
Rv0974c|M.tuberculosis_H37Rv YVDRHHARGNLTPRERIELLVDPDSPFLELSPLAAYGS-------NFQIG
MMAR_4533|M.marinum_M YVDRHHARGKLTPRERIELLVDPDSPFLELSPLAAWGS-------AFKVG
MUL_4705|M.ulcerans_Agy99 YVDRHHARGKLTPRERIELLVDPDSPFLELSPLAAWGS-------AFKVG
MAV_1090|M.avium_104 YVERHHARGKLTARERIELLVDQDSPFLELSPLAAWGS-------SFKVG
Mflv_4613|M.gilvum_PYR-GCK YVARHRSRGKMLARERVELLLDPDTAFLELCPFAAWGT-------KFPVG
MAB_2067c|M.abscessus_ATCC_199 YVARHRKRGKLLVRERIELMLDPDTAFLELCPFAAWGS-------KFPVG
TH_2815|M.thermoresistible__bu ARERHVSRGKLLPRDRVDGLLDPGSPFLEVAPLAANGMY-----DDECPG
MLBr_00731|M.leprae_Br4923 AFEKVHAKGKFTARERIYALLDDDS-FVELDALARHRSTNFGLGENRPVG
: :*.: *:*: ::* .: *:*: .:* *
MSMEG_0333|M.smegmatis_MC2_155 AAIVTGVGVVSGTECMIIAHDPTVRGGTMNPYTLRKNLRALEIAKVNRLP
Mvan_0346|M.vanbaalenii_PYR-1 AAIVTGVGVVSGVECMIIAHDPTVRGGTMNPYTLRKNLRALEIARVNRLP
Mb0999c|M.bovis_AF2122/97 ASLVTGIGAVCGVECMIVANDPTVKGGTSNPWTLRKILRANQIAFENRLP
Rv0974c|M.tuberculosis_H37Rv ASLVTGIGAVCGVECMIVANDPTVKGGTSNPWTLRKILRANQIAFENRLP
MMAR_4533|M.marinum_M ASTVTGIGVVAGVECMIVANDPTVKGGTSNPWTLKKILRANQIAFENRLP
MUL_4705|M.ulcerans_Agy99 ASTVTGIGVVAGVECMIVANDPTVKGGTSNPWTLKKILRANQIAFENRLP
MAV_1090|M.avium_104 ASTVTGIGVVEGVECMIVANDPTVKGGTSNPWTLKKILRANQIAFENRLP
Mflv_4613|M.gilvum_PYR-GCK GSVVVGIGIVEGVESMIIAHDPTVRGGASNPYTFRKVFRGMAVARENRLP
MAB_2067c|M.abscessus_ATCC_199 GSVVVGIGVVEGIESMIIAHDPTVRGGASNPYTFRKVFRGMAIARENRLP
TH_2815|M.thermoresistible__bu AGMITGIGRVSGRECMIVANDATVKGGTYYPVTVKKHLRAQEIAAQNHLP
MLBr_00731|M.leprae_Br4923 DGVVTGYGTIDGRDVCIFSQDVTVFGGSLGEVYGEKIVKVQELAIKTGRP
. :.* * : * : *.::* ** **: .* .: :* . *
MSMEG_0333|M.smegmatis_MC2_155 VLYLVESGGADLPTQSELFVHA---GRIFRDLTELSSMGIPSVAMVFGNS
Mvan_0346|M.vanbaalenii_PYR-1 VVYLVESGGADLPTQSELFVHA---GKIFHDLTELSSLGIPTISLVFGNS
Mb0999c|M.bovis_AF2122/97 VISLVESGGADLPTQKEIFIPG---GQMFRDLTRLSAAGIPTIALVFGNS
Rv0974c|M.tuberculosis_H37Rv VISLVESGGADLPTQKEIFIPG---GQMFRDLTRLSAAGIPTIALVFGNS
MMAR_4533|M.marinum_M VVSLVESGGADLPTQKEIFIPG---GRMFRDLTRLSAAGIPTIALVFGNS
MUL_4705|M.ulcerans_Agy99 VVSLVESGGADLPTQKEIFIPG---GRMFRDLTRLSAAGIPTIALVFGNS
MAV_1090|M.avium_104 VISLVESGGADLPTQKEIFIPG---GRMFRDLTRLSAAGIPTIALVFGNS
Mflv_4613|M.gilvum_PYR-GCK IINVVESGGADLPTQAEIFVPG---GQLFNDLTQHSALGLPTLALVFGNS
MAB_2067c|M.abscessus_ATCC_199 IINIVESGGADLPTQAEIFVPG---GQLFHDLTQHSAAGLPTLALVFGNS
TH_2815|M.thermoresistible__bu CIYLVDSGGAFLPRQDEVFPDRDHFGRIFYNQATLSAQGIPQIAAVMGSC
MLBr_00731|M.leprae_Br4923 LIGINDGAGARIQEGVVSLGLY---SRIFRNN-ILASGVIPQISLIMGAA
: : :..** : : .::* : :: :* :: ::* .
MSMEG_0333|M.smegmatis_MC2_155 TAGGAYVPGMCDYAVLVDKQAKVFLGGPPLVKMATGEDADDESLGGADMH
Mvan_0346|M.vanbaalenii_PYR-1 TAGGAYVPGMCDYAVLVDKQAKVFLGGPPLVKMATGEESDDESLGGADMH
Mb0999c|M.bovis_AF2122/97 TAGGAYVPGMSDHVVMIKERSKVFLAGPPLVKMATGEESDDESLGGAEMH
Rv0974c|M.tuberculosis_H37Rv TAGGAYVPGMSDHVVMIKERSKVFLAGPPLVKMATGEESDDESLGGAEMH
MMAR_4533|M.marinum_M TAGGAYVPGMSDHVVMIKERSKVFLAGPPLVKMATGEESDDESLGGAEMH
MUL_4705|M.ulcerans_Agy99 TAGGAYVPGMSDHVVMIKERSKVFLAGPPLVKMATGEESDDESLGGAEMH
MAV_1090|M.avium_104 TAGGAYVPGMSDHVVMIKERSKVFLAGPPLVKMATGEESDDESLGGAEMH
Mflv_4613|M.gilvum_PYR-GCK TAGGAYIPGMCDYVVMVRNRSKVFLGGPPLVKMATGEVSDDESLGGADMH
MAB_2067c|M.abscessus_ATCC_199 TAGGAYVPGMCDYVVMVRNRSKVFLGGPPLVKMATGEVSDDESLGGADMH
TH_2815|M.thermoresistible__bu TAGGAYVPAMSDEAVIVRNQGTIFLGGPPLVRAATGEVVTAEELGGGDLH
MLBr_00731|M.leprae_Br4923 AGGHVYSPALTDFVVMVDQTSQMFITGPDVIKTVTGEDVTMEELGGAHTH
:.* .* *.: * .*:: : . :*: ** ::: .*** *.***.. *
MSMEG_0333|M.smegmatis_MC2_155 TRVSGLGDYFAVDEVDAIRIGRDIVAHLNWRKLGPGPSRPADEPLYDSEE
Mvan_0346|M.vanbaalenii_PYR-1 TRVSGLGDYFAVDETDAIRIGRDIVAHLNWHKLGPGPTQPADEPLYDVEE
Mb0999c|M.bovis_AF2122/97 ARISGLADYFALDELDAIRIGRRIVARLNWIKQGPAP-APVTEPLFDAEE
Rv0974c|M.tuberculosis_H37Rv ARISGLADYFALDELDAIRIGRRIVARLNWIKQGPAP-APVTEPLFDAEE
MMAR_4533|M.marinum_M ARISGLADYFAVDELDAIRIGRRIVARLNWVKQGPAP-APVTEPLFDAEE
MUL_4705|M.ulcerans_Agy99 ARISGLADYFAVDEFDAIRIGRRIVARLNWVKQGPAP-APVTEPLFDAEE
MAV_1090|M.avium_104 ARVSGLADYFAVDELDAIRIGRRIVARLNWVKQGPTP-KPVTEPLFDTEE
Mflv_4613|M.gilvum_PYR-GCK ARTSGLADYMAEDEQDAIRIGRRIMARLNWRKLGPGPTQPPTPPLRDPDQ
MAB_2067c|M.abscessus_ATCC_199 ARTSGLADYMAEDEQDAIRIGRRIMARLNWRKSGPGPTTPPHPPVHDPDQ
TH_2815|M.thermoresistible__bu AKTSGVTDHLAHDDRDALRIVRRIVATLGPRHPSPWEIAPTVDPVADQTE
MLBr_00731|M.leprae_Br4923 MAKSGTAHYVASGEQDAFDWVRDVLSYLPSNNFTDAPRYSKPVPHGSIED
** .:.* .: **: * ::: * : . * . :
MSMEG_0333|M.smegmatis_MC2_155 --------LLGIASGDSRVPFDPREVIARIVDGSRFSEYKPRWGTSLATG
Mvan_0346|M.vanbaalenii_PYR-1 --------LLGIASGDSRVPFDPREVIARVADGSRFSEYKPRWGTSLATG
Mb0999c|M.bovis_AF2122/97 --------LIGIVPPDLRIPFDPREVIARIVDGSEFDEFKPLYGSSLVTG
Rv0974c|M.tuberculosis_H37Rv --------LIGIVPPDLRIPFDPREVIARIVDGSEFDEFKPLYGSSLVTG
MMAR_4533|M.marinum_M --------LIGIVPSDLRVPFDPREVIARIVDGSEFDEFKPLYGSSLVTG
MUL_4705|M.ulcerans_Agy99 --------LIGIVPSDLRVPFEPREVIARIVDGSEFDEFKPLYGSSLVTG
MAV_1090|M.avium_104 --------LIGIVPADLRIPFDPREVIARIVDGSEFDEFKPLYGSSLVTG
Mflv_4613|M.gilvum_PYR-GCK --------LLGIASVDPKVPFDPRDVIARIVDGSTFDEFKPLYGTSLVTG
MAB_2067c|M.abscessus_ATCC_199 --------LLGIASVDPKVPFDPRDVIARVVDGSTFDEFKPLYGTSLVTG
TH_2815|M.thermoresistible__bu --------LYDVVPVDPRIPYDVHEVVDRIVDGGDFAEFKAEYGATLVTG
MLBr_00731|M.leprae_Br4923 NLTAKDLELDTLIPDSPNQPYDMHEVVTRLLDEEEFLEVQAGYATNIVVG
* : . . . *:: ::*: *: * * * :. :.:.:..*
MSMEG_0333|M.smegmatis_MC2_155 WASIHGFPVGILANTR----GVLFSEESKKATEFILLANQTDTPLIFLQN
Mvan_0346|M.vanbaalenii_PYR-1 WASIHGFPVGILANTR----GVLFSEESKKATEFILLANQTDTPLIFLQN
Mb0999c|M.bovis_AF2122/97 WARLHGYPLGILANAR----GVLFSEESQKATQFIQLANRADTPLLFLHN
Rv0974c|M.tuberculosis_H37Rv WARLHGYPLGILANAR----GVLFSEESQKATQFIQLANRADTPLLFLHN
MMAR_4533|M.marinum_M WARLHGYPVGILANAR----GVLFSEESQKATQFIQLANRADTPLLFLHN
MUL_4705|M.ulcerans_Agy99 WARLHGYPVGILANAR----GVLFSEESQKATQFIQLANRADTPLLFLHN
MAV_1090|M.avium_104 WARLHGYPIGILANAR----GVLFSEESQKATQFIQLANRSNTPLLFLHN
Mflv_4613|M.gilvum_PYR-GCK WASIHGYPVGILANAR----GILFSEEAEKAAQFIQLANQIDTPLVFLQN
MAB_2067c|M.abscessus_ATCC_199 WASIHGFPVGILANAR----GILFSEEAEKGSQFIQLANQIDTPLIFLQN
TH_2815|M.thermoresistible__bu FARIHGHPVGIIANN-----GVLFSESALKGAHFIELCDKRKIPLLFLQN
MLBr_00731|M.leprae_Br4923 LGRIDDRPVGIVANQPIQFAGCLDINASEKAARFVRVCDCFNIPIVMLVD
. :.. *:**:** * * : : *.:.*: :.: . *:::* :
MSMEG_0333|M.smegmatis_MC2_155 VTGFMVGTEYEQGGIIKDGAKMINAVTNSTVPHITLNMGGSFGAGNYGMS
Mvan_0346|M.vanbaalenii_PYR-1 VTGFMVGAEYEQGGIIKDGAKMINAVTNSTVPHITINMGGSFGAGNYGMS
Mb0999c|M.bovis_AF2122/97 TTGYMVGKDYEEGGMIKHGSMMINAVSNSTVPHISLLIGASYGAGHYGMC
Rv0974c|M.tuberculosis_H37Rv TTGYMVGKDYEEGGMIKHGSMMINAVSNSTVPHISLLIGASYGAGHYGMC
MMAR_4533|M.marinum_M TTGYMVGKDYEEGGMIKHGSMMINAVSNSKVPHISLLLGASYGAGHYGMC
MUL_4705|M.ulcerans_Agy99 TTGYMVGKDYEEGGMIKHASMMINAVSNSKVPHISLLLGASYGAGHYGMC
MAV_1090|M.avium_104 TTGYMVGKDYEEGGMIKHGSMMINAVSNSTVPHISLLIGASYGAGHYGMC
Mflv_4613|M.gilvum_PYR-GCK TTGYMVGAEYEQGGIIKDGAKMINAVSNSAVPHLTIVMGASYGAGNYGMC
MAB_2067c|M.abscessus_ATCC_199 TTGFMVGAEYEQGGIIKDGAKMINAVSNSTVPHVTITMGASYGAGNYGMC
TH_2815|M.thermoresistible__bu ISGFMVGREYEAGGIAKHGAKMVTAVACARVPKLTVVIGGSYGAGNYSMC
MLBr_00731|M.leprae_Br4923 VPGFLPGTEQEYDGIIRRGAKLLFAYGEATVPKITVITRKAYGGAYCVMG
.*:: * : * .*: : .: :: * : **:::: ::*.. *
MSMEG_0333|M.smegmatis_MC2_155 GRAYDPRFMFSWVNAKLAVMGAQQLAGVLSIVGKAAAQAAGREFDHEADA
Mvan_0346|M.vanbaalenii_PYR-1 GRAYDPRFMFSWVNAKLAVMGAAQLAGVLSIVGKAAAAAAGREFDREADA
Mb0999c|M.bovis_AF2122/97 GRAYDPRFLFAWPSAKSAVMGGAQLSGVLSIVARAAAEARGQQVDEAADA
Rv0974c|M.tuberculosis_H37Rv GRAYDPRFLFAWPSAKSAVMGGAQLSGVLSIVARAAAEARGQQVDEAADA
MMAR_4533|M.marinum_M GRAYDPRFLFAWPSAKSAVMGGAQLSGVLSIVARAAAEARGQQVDEAADA
MUL_4705|M.ulcerans_Agy99 GRAYDPRFLFAWPSAKSAVMGGAQLSGVLSIVARAAAEARGQQVDEAADA
MAV_1090|M.avium_104 GRAYDPRFLFAWPTSKSAVMGGAQLAGVLSIVARAAAEARGQQFDEQADA
Mflv_4613|M.gilvum_PYR-GCK GRSYSPRFLFTWPNSRSAVMGPAQLAGVLSIVARESAADRGLPFDEEADA
MAB_2067c|M.abscessus_ATCC_199 GRSYSPRFLFAWPNSKSAVMGPAQLAGVLSIVARESAADRGLPFDEEADA
TH_2815|M.thermoresistible__bu GRAYSPRFLWMWPNARISVMGGEQAASVLATVR--------GDMTPEEEE
MLBr_00731|M.leprae_Br4923 SKNMGCDVNLAWPTAQIAVMGASGAVGFVYRKELAQAAKNGANVDELRLQ
.: . . * .:: :*** ..: .
MSMEG_0333|M.smegmatis_MC2_155 KRTKEIEEQIDRESHAFFNSARIYDDGVIDPRDTRSVLGIALSAAHSNVV
Mvan_0346|M.vanbaalenii_PYR-1 KRTAEIEAQIEKESHSFFVSARIYDDGVIDPRDTRSVLGMALSAAHSNVV
Mb0999c|M.bovis_AF2122/97 AMRAAVEGQIEAESLPLVLSGMLYDDGVIDPRDTRTVLGMCLSAIANGPI
Rv0974c|M.tuberculosis_H37Rv AMRAAVEGQIEAESLPLVLSGMLYDDGVIDPRDTRTVLGMCLSAIANGPI
MMAR_4533|M.marinum_M AMRAAVEGQIEAESLPLVLSGMLYDDGVIDPRDTRTVLGMCLSAIANGPI
MUL_4705|M.ulcerans_Agy99 AMRAAVDGQIEAESLPLVLSGMLYDDGVIDPRDTRTVLGMCLSAIANGPI
MAV_1090|M.avium_104 AMRAAVEGQIEAESLPLVLSGMLYDDGVIDPRDTRTVLGMCLSAIANGPI
Mflv_4613|M.gilvum_PYR-GCK QMRAAVEGQIERESLALANSGRLYDDGIIDPRDTRTVLGFCLSVVHNSEI
MAB_2067c|M.abscessus_ATCC_199 QMRAAVEEQIERESVALANSGRLYDDGIIDPRDTRTVLGFCLSVIHNGEV
TH_2815|M.thermoresistible__bu AFKVPIRQQYEDQGNPYYSTARLWDDGVIDPADTRTVVGLALSIVGQAPL
MLBr_00731|M.leprae_Br4923 LQQEYEDTLVN----PYIAAERGYVDAVIPPSHTRGYIATALHLLERKIA
: . : : *.:* * .** :. .*
MSMEG_0333|M.smegmatis_MC2_155 A-GRRGFGVFRM
Mvan_0346|M.vanbaalenii_PYR-1 E-GRRGFGVFRM
Mb0999c|M.bovis_AF2122/97 K-GTSNFGVFRM
Rv0974c|M.tuberculosis_H37Rv K-GTSNFGVFRM
MMAR_4533|M.marinum_M K-GTSNFGVFRM
MUL_4705|M.ulcerans_Agy99 K-GTSNFGVFRM
MAV_1090|M.avium_104 E-GTSNFGVFRM
Mflv_4613|M.gilvum_PYR-GCK R-GQRGYGVFRM
MAB_2067c|M.abscessus_ATCC_199 R-GQRGYGVFRM
TH_2815|M.thermoresistible__bu E-PVS-YGVFRM
MLBr_00731|M.leprae_Br4923 HLPPKKHGNIPL
.* : :