For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKTVYYTASSLDGYIVDENQSLDWLTSRDITPDGPFGYEQFIETIGVLVMGASTYEWVVEHGDWSYDQPA WVLTHRPEIAAESHPMQVFSGDVAELHPELVAAAGGKDVWVVGGGDVAAQFVAADLIDEIIVSYAPCTLG VGSRVLPMRSEWVLDDCARNGDFVCARWKRPVLHT
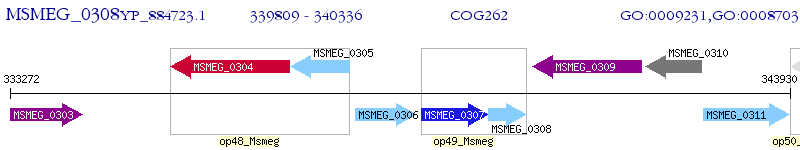
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0308 | - | - | 100% (175) | dihydrofolate reductase |
| M. smegmatis MC2 155 | MSMEG_5219 | - | 2e-06 | 25.16% (159) | riboflavin biosynthesis protein RibD domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_3449 | - | 1e-05 | 31.09% (119) | DNA-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0431 | - | 2e-68 | 66.86% (172) | bifunctional deaminase-reductase-like protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2852 | - | 3e-21 | 38.71% (155) | hypothetical protein MMAR_2852 |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_1907 | - | 4e-71 | 67.65% (170) | dihydrofolate reductase |
| M. ulcerans Agy99 | MUL_2905 | - | 4e-21 | 38.71% (155) | hypothetical protein MUL_2905 |
| M. vanbaalenii PYR-1 | Mvan_0237 | - | 3e-71 | 70.93% (172) | bifunctional deaminase-reductase-like protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0431|M.gilvum_PYR-GCK --MSTIYYTASSLDGYIVDDHGSLDWLTSRAIDADGPFGIEPFMADIGSL
Mvan_0237|M.vanbaalenii_PYR-1 --MSTVYYTASSLDGYIVDENKSLDWLTSREIAADGPFAIDPFMETVGSL
MSMEG_0308|M.smegmatis_MC2_155 --MKTVYYTASSLDGYIVDENQSLDWLTSRDITPDGPFGYEQFIETIGVL
TH_1907|M.thermoresistible__bu LIMATVYFTAASLDGFIVDDHDSLDWLVTRDIDPDGPFGYRAFADSVGAV
MMAR_2852|M.marinum_M -MGQIVFDTATTLNGWIADENNSLAWLFAVDQGGLPESGLVPTEATV--M
MUL_2905|M.ulcerans_Agy99 -MGQIVFDTATTLNGWIADENNSLAWLFAVDQGGLPESGLVPTEATV--M
:: **::*:*:*.*:: ** ** : . : :
Mflv_0431|M.gilvum_PYR-GCK VMGADTYEWILRNH--------PGEWMYTQPSWVVTHRPDIVAADHPVRT
Mvan_0237|M.vanbaalenii_PYR-1 VMGADTYEWILENH--------PGEWMYPQPTWVLTHRPGIIADGHDVQT
MSMEG_0308|M.smegmatis_MC2_155 VMGASTYEWVVEH----------GDWSYDQPAWVLTHRPEIAAESHPMQV
TH_1907|M.thermoresistible__bu VMGAATYEWVLRNQ--------PGEWMHEQPSWVLTHRPQIIAEAHPVRT
MMAR_2852|M.marinum_M VEGSTTYQWVIETESILDHPEKWQQFYGDKPTFVFTTRELPVPAGADVRF
MUL_2905|M.ulcerans_Agy99 VEGSTTYQWVIETESILDHPEKWQQFYGDKPTFVFTTRELPVPAGADVRF
* *: **:*::. :: :*::*.* * . ::
Mflv_0431|M.gilvum_PYR-GCK FAGDVVDLHPTLLDAAAGRNVWVVGGGDLARQFAAAGLIDEMIVSYAPCT
Mvan_0237|M.vanbaalenii_PYR-1 FAGDVADLHPVLEEAAAGKNVWLVGGGDVAAQFVAAGLVDEMIVSYAPCT
MSMEG_0308|M.smegmatis_MC2_155 FSGDVAELHPELVAAAGGKDVWVVGGGDVAAQFVAADLIDEIIVSYAPCT
TH_1907|M.thermoresistible__bu FSGDVAELHPELVRAAGDKDVWVVGGGEVAAQFVAAGLVDDVIIGYAPCT
MMAR_2852|M.marinum_M VSGSVLDVLPTIEEAAGAGDIWVVGGGDLAGQFLDAGALDEIRLSVAPVA
MUL_2905|M.ulcerans_Agy99 VSGSVLDVLPTIEEAAGAGDIWVVGGGDLAGQFLDAGALDEIRSSVAPVA
.:*.* :: * : **. ::*:****::* ** *. :*:: . ** :
Mflv_0431|M.gilvum_PYR-GCK LGAGSPVLP---MRSEWTLQTCAANADFVCAHWRRV----
Mvan_0237|M.vanbaalenii_PYR-1 LGAGSRVLP---MRSEWALVSSAVNADFVCAHWRRA----
MSMEG_0308|M.smegmatis_MC2_155 LGVGSRVLP---MRSEWVLDDCARNGDFVCARWKRPVLHT
TH_1907|M.thermoresistible__bu LGAGAPVLP---QRSEWTLVDSGVNGEFLCARWSRSAGS-
MMAR_2852|M.marinum_M LAGGAPLLPRCIGADRLRLVSAAAHGQFARLVYTVTSPE-
MUL_2905|M.ulcerans_Agy99 LAGGAPLLPRCIGADRLRLVSAAAHGQFARLVYTVTSPE-
*. *: :** .. * .. :.:* :