For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GVITDDVTIAQQALTHYDVSDNASLRLLNLSENATYLVEDGEHQSILRVHRQDYHQPHEIESELDWLAAL RTDSDVTVPTVVPARDGRRVVTVDPADSDAVPRHVVHFEMVGGAEPDEESLTLDDFQTLGRITASLHEHS QRWTRPAGFGRFSWDWEHCLGDTPRWGRWQDAEGVGASETALLTRAQDLLHRKLEEYGSGPDRYGLIHAD LRLANLLVDSSTPQRTITVIDFDDCGFGWYFYDFGTAVSFIEHDPRLGEWQESWVAGYRSRRELPAADEA MLPSFVFLRRLLLLAWMGSHTHSRESATKAISYAAGSCALAERYLSSDGLRLT*
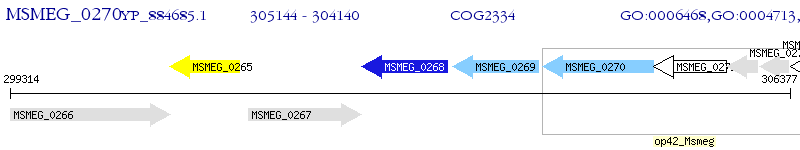
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0270 | - | - | 100% (334) | aminoglycoside phosphotransferase |
| M. smegmatis MC2 155 | MSMEG_0780 | - | 6e-72 | 43.60% (328) | phosphotransferase enzyme family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1236 | - | 1e-130 | 67.07% (331) | aminoglycoside phosphotransferase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0951 | - | 1e-68 | 41.41% (326) | putative aminoglycoside phosphotransferase |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_1675 | - | 1e-72 | 43.38% (325) | phosphotransferase enzyme family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5572 | - | 1e-129 | 67.17% (332) | aminoglycoside phosphotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1236|M.gilvum_PYR-GCK MTAD-FTDDQTVAEKALAAFDLPQGSALRLLNLSENATYAVEEPGCGHRS
Mvan_5572|M.vanbaalenii_PYR-1 MSGDDVNDDRAVAEKALEAFDLPQGSTLRLLNLSENATYAVEEPGCGHRS
MSMEG_0270|M.smegmatis_MC2_155 --MGVITDDVTIAQQALTHYDVSDNASLRLLNLSENATYLVEDG--EHQS
MAB_0951|M.abscessus_ATCC_1997 -MPGLPPNHSAFARAALTSYGRDPHTPLRLLSLSENATYLVDDQ---RPM
TH_1675|M.thermoresistible__bu -MSGLPRDHERHARTVLPAYGRDPDTPLRLLSVSENAIYLVDDP---DPM
. :. *. .* :. :.****.:**** * *::
Mflv_1236|M.gilvum_PYR-GCK ILRVHRKDYHRVDQIESELMWLDALRRDSDVTVPTVIPAQDGRRVVTVEH
Mvan_5572|M.vanbaalenii_PYR-1 ILRVHRQNYHRVDQIESELLWLDALRRDSDVTVPTVLPAHDGRRVVTVAH
MSMEG_0270|M.smegmatis_MC2_155 ILRVHRQDYHQPHEIESELDWLAALRTDSDVTVPTVVPARDGRRVVTVDP
MAB_0951|M.abscessus_ATCC_1997 VLRAHRPGYHSLPAIRSELAWMSALRAETSVVTPELVRARNGSEVIAVGV
TH_1675|M.thermoresistible__bu VLRVHRPGYHSPAAIRSELGWMTALREQTTVATPRALRTVDGRDVVAAQP
:**.** .** *.*** *: *** :: *..* : : :* *::.
Mflv_1236|M.gilvum_PYR-GCK DGDE---RYVVHFEMVPGAEPDEDTVTSTDFHTLGCITAALHDHARRWQR
Mvan_5572|M.vanbaalenii_PYR-1 DGAD---RHVVHFEMVPGAEPDENALTSTDFHTLGRITAALHDHARSWER
MSMEG_0270|M.smegmatis_MC2_155 ADSDAVPRHVVHFEMVGGAEPDEESLTLDDFQTLGRITASLHEHSQRWTR
MAB_0951|M.abscessus_ATCC_1997 DG---ITLHVDAMGFVPGCTAEE-TSGVVGYGKLGQLTAHMHHHAQGWRV
TH_1675|M.thermoresistible__bu DGSRGSPVYVDAVTFIPGCTAEE-QPDAVGFEDLGRITAVMHRHVQSWSP
. :* . :: *. .:* .: ** :** :* * : *
Mflv_1236|M.gilvum_PYR-GCK PSGFSRFAWDWEHSLGGTPRWGRWRDAVGVGEHEADVLTRAEALLHRRLT
Mvan_5572|M.vanbaalenii_PYR-1 PAAFSRFAWDWENSLGGSPRWGRWRDAIGVGAQEAEVLERAERLLHRRLA
MSMEG_0270|M.smegmatis_MC2_155 PAGFGRFSWDWEHCLGDTPRWGRWQDAEGVGASETALLTRAQDLLHRKLE
MAB_0951|M.abscessus_ATCC_1997 PPGFTRFRWDLDSMFGPQARWGDWRQAPGLTDADREAVDAAVRDIIRRLS
TH_1675|M.thermoresistible__bu PAGFTRFRWDIDTTIGAHPRWGSWRHAAGLTPRDIDVVERAAAAVTARLT
*..* ** ** : :* .*** *:.* *: : : * : :*
Mflv_1236|M.gilvum_PYR-GCK DYGTGPDAFGLVHADLRLANLLVDDDT----ITVIDFDDCGMGWYFYDFG
Mvan_5572|M.vanbaalenii_PYR-1 EYGTGPDTFGLVHADLRLANLLVDGDT----ITVIDFDDCGFGWYFYDFG
MSMEG_0270|M.smegmatis_MC2_155 EYGSGPDRYGLIHADLRLANLLVDSSTPQRTITVIDFDDCGFGWYFYDFG
MAB_0951|M.abscessus_ATCC_1997 EYGCTPDRFGLIHADMRLSNLMVDPGRPGSDITVIDFDDCGWSWFMADLG
TH_1675|M.thermoresistible__bu EFGYAPDKFGLIHGDLRMANLMIDPADPGAPITVIDFDDSGWSWYLADLG
::* ** :**:*.*:*::**::* ********.* .*:: *:*
Mflv_1236|M.gilvum_PYR-GCK TAVSFFEDHPSVPEWQDAWVSGYRSRRPISAADEDMLASFILLRRLLLLA
Mvan_5572|M.vanbaalenii_PYR-1 TAVSFFEDHPSVPEWQEAWVSGYRTRRTLAASDEDMLASFILLRRLLLLA
MSMEG_0270|M.smegmatis_MC2_155 TAVSFIEHDPRLGEWQESWVAGYRSRRELPAADEAMLPSFVFLRRLLLLA
MAB_0951|M.abscessus_ATCC_1997 AVVSWVEDTPMAEAAVADWLDGYQRVRRLSGDDLGMIPAFVMLRRVMLTA
TH_1675|M.thermoresistible__bu AVVSFIEHTPLAESIIADWLCGYREVCELPQADLEMVPTFVMLRRLMLTG
:.**:.*. * *: **: :. * *:.:*::***::* .
Mflv_1236|M.gilvum_PYR-GCK WMGTHSHSKESQAISVTYAAGSCTLAEHYLSSD--------GHRIF
Mvan_5572|M.vanbaalenii_PYR-1 WMGTHSHSKESQAISVTYAAGSCKLAEQYLSSD--------GHRLF
MSMEG_0270|M.smegmatis_MC2_155 WMGSHTHSRESATKAISYAAGSCALAERYLSSD--------GLRLT
MAB_0951|M.abscessus_ATCC_1997 WIASHADADAAIRVSDGFGARTARLAERYLTDRTWLREAVFGRRVG
TH_1675|M.thermoresistible__bu WIASHPDADAAITYRDGYPEGTVQLAHRYLGDRNWLRDAVFSTRV-
*:.:*..: : : : **.:** . . *: