For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AGAQDFVPHTADLAELAAAAGECRGCGLYRDATQAVFGAGGRSARIMMIGEQPGDKEDLAGLPFVGPAGR LLDRALEAADIDRDALYVTNAVKHFKFTRAAGGKRRIHKTPSRTEVVACRPWLIAEMTSVEPDVVVLLGA TAAKALLGNDFRVTQHRGEVLHVDDVPGDPALVATVHPSSLLRGPKEERESAFAGLVDDLRVAADVRP*
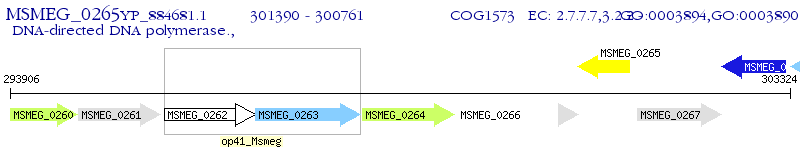
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0265 | - | - | 100% (209) | uracil DNA glycosylase superfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_4094 | - | 3e-61 | 57.89% (209) | uracil DNA glycosylase superfamily protein |
| M. thermoresistible (build 8) | TH_1350 | - | 1e-70 | 65.88% (211) | uracil DNA glycosylase superfamily protein |
| M. ulcerans Agy99 | MUL_0027 | - | 3e-08 | 24.16% (178) | glycosylase |
| M. vanbaalenii PYR-1 | Mvan_3202 | - | 2e-72 | 64.15% (212) | phage SPO1 DNA polymerase-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
TH_1350|M.thermoresistible__bu ----MAVTGAARFVPATRDLGELAEAVHACKGCD--LYVDATQAVFGAGP
Mvan_3202|M.vanbaalenii_PYR-1 ----MTIVGAAEFVPQTRELSTLAAAAQSCRGCD--LFENASRAVFGSGP
MSMEG_0265|M.smegmatis_MC2_155 ------MAGAQDFVPHTADLAELAAAAGECRGCG--LYRDATQAVFGAGG
MAV_4094|M.avium_104 MTATSKAPGADRYLPEQRDIEALKHAAETCRGCS--LFADATQTVFGNGH
MUL_0027|M.ulcerans_Agy99 -MGPSVTEDYDSSASRRLRKIALDAEIAGCRRCEGMNIAPATMSAPGYGC
. . * *: * *: :. * *
TH_1350|M.thermoresistible__bu GTAPMVMVGEQPGDREDTAGQPFVGPAGRLLQRALDAAGIDRAEVYVTNA
Mvan_3202|M.vanbaalenii_PYR-1 VGARMMLVGEQPGDREDVAGEPFVGPAGRLLDKALDEAGLPRAPVYLTNA
MSMEG_0265|M.smegmatis_MC2_155 RSARIMMIGEQPGDKEDLAGLPFVGPAGRLLDRALEAADIDRDALYVTNA
MAV_4094|M.avium_104 PGAPIMLVGEQPGDQEDRAGAPFVGPAGRLLARALHDAGIDPGLTYQTNA
MUL_0027|M.ulcerans_Agy99 VSSPVALIGQSLCEKCMESQVPFTGGSGELIDESIQRAGREKEDVFISDA
: : ::*:. :: : **.* :*.*: .::. *. : ::*
TH_1350|M.thermoresistible__bu VKHFTFT---RGRRLIHKTPSRSDVVACRPWLIAELDSVRPEVVVLLGAT
Mvan_3202|M.vanbaalenii_PYR-1 VKHFKFVPAERGTRRIHKTPSRTEVVACRPWLIAEMQAVEPAVVVLLGAT
MSMEG_0265|M.smegmatis_MC2_155 VKHFKFTRAAGGKRRIHKTPSRTEVVACRPWLIAEMTSVEPDVVVLLGAT
MAV_4094|M.avium_104 VKHFKFTR-KDGKRRIHQKPGRTEIVACRPWLIAEIEAVHPRVIVCLGAT
MUL_0027|M.ulcerans_Agy99 VHCHPPK---------NRSSHQHEIVDCSPYLHRELEIVRPRLVVALGRD
*: . ::.. : ::* * *:* *: *.* ::* **
TH_1350|M.thermoresistible__bu AAKSLLGPDFRLTAHRGEVLRLPAGDATV-GLGVDPLVVVTVHPSAVLRG
Mvan_3202|M.vanbaalenii_PYR-1 AAKSLLGNAFRLTAHRGEVLSLPAETKPV-G-GRDAKVVVTVHPSSVLRG
MSMEG_0265|M.smegmatis_MC2_155 AAKALLGNDFRVTQHRGEVLHVDDVPG-------DPALVATVHPSSLLRG
MAV_4094|M.avium_104 AAQSLLGTSFRVSAQRGQPLKLPASPEVIPDVAPEPVLVATVHPSSVLRD
MUL_0027|M.ulcerans_Agy99 GER-VLTFFYPSARICEELFEIPRGRLPK----GVPYIFHVKHPSWIQRQ
. : :* : : : : : . :. . *** : *
TH_1350|M.thermoresistible__bu RPGDRAEAFDALVADLEVAAGLMGS-------------------------
Mvan_3202|M.vanbaalenii_PYR-1 PSQAREESFRSLVSDLRFAGELLDA-------------------------
MSMEG_0265|M.smegmatis_MC2_155 PKEERESAFAGLVDDLRVAADVRP--------------------------
MAV_4094|M.avium_104 RSERHDEVYRLFVDDLRSARSALG--------------------------
MUL_0027|M.ulcerans_Agy99 HDDNLEANYGRTCPRRSGGASTAKARKRRARRDVASADVQFLDGYPVLSH
: .
TH_1350|M.thermoresistible__bu ----------------
Mvan_3202|M.vanbaalenii_PYR-1 ----------------
MSMEG_0265|M.smegmatis_MC2_155 ----------------
MAV_4094|M.avium_104 ----------------
MUL_0027|M.ulcerans_Agy99 AVPCRPAWCIPRVPES