For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDRTFGLATRAIHAGQRPDAATGARVTPIYANASFVFNDTDDAANLFALQKYGNIYSRIGNPTVAAFEEC IASLENGLGAVAMASGLSAQFAVFAALAGAGDHIVSAASLYGGTVTQLDVSLRRFGIDTTFVGGTDPADY AAAVTDRTRLIYAEVISNPSGEIADLAGLAEVARDAGVPLIVDSTVATPALCRPIEHGADIVVHSATKFI GGHGTTLGGVAVDAGRFDWGAGKFPQMTEPVASYGGLSWWGNFGEFGFLTKLRSEQLRDLGATLAPQSAF QLLQGVQTLPQRMAAHIANARAVAEWLEADPRVTYVRWAGLPSHPHHDRARHYLPDGPGAVFAFGVKGGR AAGEKFINSVQLASHLANIGDVRTLVVHPASTTHRQLTDEQLTAGGVGPELIRISVGIEDVEDIIWDLDQ ALTQAEKADD*
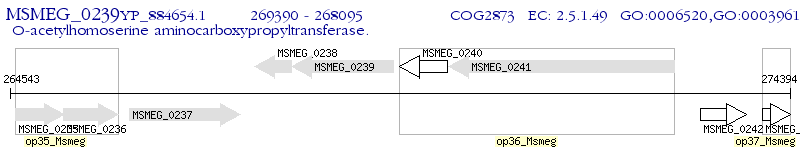
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0239 | - | - | 100% (431) | O-acetylhomoserine/O-acetylserine sulfhydrylase |
| M. smegmatis MC2 155 | MSMEG_1652 | - | e-117 | 51.44% (418) | O-acetylhomoserine aminocarboxypropyltransferase |
| M. smegmatis MC2 155 | MSMEG_5265 | - | 2e-61 | 35.01% (417) | cystathionine gamma-synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3372 | metC | 1e-113 | 49.76% (420) | O-acetylhomoserine aminocarboxypropyltransferase |
| M. gilvum PYR-GCK | Mflv_4875 | - | 1e-114 | 49.65% (429) | O-acetylhomoserine aminocarboxypropyltransferase |
| M. tuberculosis H37Rv | Rv3340 | metC | 1e-113 | 49.76% (420) | O-acetylhomoserine aminocarboxypropyltransferase |
| M. leprae Br4923 | MLBr_02394 | metB | 1e-64 | 36.21% (417) | cystathionine gamma-synthase |
| M. abscessus ATCC 19977 | MAB_3687 | - | 1e-115 | 50.48% (420) | O-acetylhomoserine aminocarboxypropyltransferase |
| M. marinum M | MMAR_1185 | metC | 1e-112 | 49.05% (420) | O-acetylhomoserine sulfhydrylase MetC |
| M. avium 104 | MAV_4315 | - | 1e-114 | 49.76% (420) | O-acetylhomoserine aminocarboxypropyltransferase |
| M. thermoresistible (build 8) | TH_1970 | metC | 1e-115 | 51.31% (419) | PROBABLE O-ACETYLHOMOSERINE SULFHYDRYLASE METC (HOMOCYSTEINE |
| M. ulcerans Agy99 | MUL_1438 | metC | 1e-111 | 48.81% (420) | O-acetylhomoserine aminocarboxypropyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1554 | - | 1e-113 | 49.18% (425) | O-acetylhomoserine aminocarboxypropyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4875|M.gilvum_PYR-GCK MTD---PDLT--------AGWAFETKQVHAGQTPDSATNARALPIYQTTS
Mvan_1554|M.vanbaalenii_PYR-1 MTD---TTAAN-------ASWAFETKQVHAGQTPDIATNARALPIYQTTS
TH_1970|M.thermoresistible__bu MTN---SEQDDPT-----AKWSFETKQVHAGQTPDPATNARALPIYQTTS
Mb3372|M.bovis_AF2122/97 MSADSNSTDADPT-----AHWSFETKQIHAGQHPDPTTNARALPIYATTS
Rv3340|M.tuberculosis_H37Rv MSADSNSTDADPT-----AHWSFETKQIHAGQHPDPTTNARALPIYATTS
MMAR_1185|M.marinum_M MSAENTSTDADPT-----AHWSFETKQIHAGQQPDSATNARALPIYQTTS
MUL_1438|M.ulcerans_Agy99 MSAENTSTDADPT-----AHWSFETKQIHAGQQPDSATNARALPIYQTTS
MAV_4315|M.avium_104 MSSENTDHDTDPS-----AHWSFETKQVHAGQHPDSATNARALPIYQTTS
MAB_3687|M.abscessus_ATCC_1997 MTDIDPTTADSRSDKPRSAQWAFETKQIHAGQSADAATNARALPIYQTTS
MSMEG_0239|M.smegmatis_MC2_155 MTD---------------RTFGLATRAIHAGQRPDAATGARVTPIYANAS
MLBr_02394|M.leprae_Br4923 MSEDYRGHHG---------ITGLATKAIHAGYRPDPATGAVNVPIYASST
*: .: *: :*** .* :*.* *** .::
Mflv_4875|M.gilvum_PYR-GCK YIFDSTDHAAALFGLAEPGNIYTRIMNPTQDVVEQRIAALEGGVAALFLS
Mvan_1554|M.vanbaalenii_PYR-1 YTFNSTDHAAALFGLAEPGNIYTRIMNPTTDVVEQRIAALEGGVAALFLA
TH_1970|M.thermoresistible__bu YTFDSTEHAAALFGLSEPGNIYTRIMNPTQDVVEKRIAALEGGVAALFLA
Mb3372|M.bovis_AF2122/97 YTFDDTAHAAALFGLEIPGNIYTRIGNPTTDVVEQRIAALEGGVAALFLS
Rv3340|M.tuberculosis_H37Rv YTFDDTAHAAALFGLEIPGNIYTRIGNPTTDVVEQRIAALEGGVAALFLS
MMAR_1185|M.marinum_M YTFENTAHAAALFGLEVPGNIYTRLGNPTTDVVEQRIAALEGGVAALFLS
MUL_1438|M.ulcerans_Agy99 YTFENTAHAAALFGLEVPGNIYTRLGNPTTDVVEQRIAALEGGVAALFLS
MAV_4315|M.avium_104 YTFDDTTHAAALFGLEVPGNIYTRIGNPTTDVVEQRIAALEGGVAALFLS
MAB_3687|M.abscessus_ATCC_1997 YVFNDTAHAAALFGLSEEGNIYTRIGNPTQDVVEQRIAALEGGVAALLLA
MSMEG_0239|M.smegmatis_MC2_155 FVFNDTDDAANLFALQKYGNIYSRIGNPTVAAFEECIASLENGLGAVAMA
MLBr_02394|M.leprae_Br4923 FAQDG-------VGELRGGFEYARTGNPMRAALEASLATVEEGVFARAFS
: :. .. * *:* ** ..* :*::* *: * ::
Mflv_4875|M.gilvum_PYR-GCK SGQAAETLAILNIAGAGDHIVSSPRLYGGTYNLFHYTLPKLGIEVSFVED
Mvan_1554|M.vanbaalenii_PYR-1 SGQAAETFAILNLANHGDHIVSSPRLYGGTYNLFHYTLPKLGIEVSFVEN
TH_1970|M.thermoresistible__bu SGQAAETFAILNLANAGDHIVASPRLYGGTYNLFQHTLPKLGIETTFVEN
Mb3372|M.bovis_AF2122/97 SGQAAETFAILNLAGAGDHIVSSPRLYGGTYNLFHYSLAKLGIEVSFVDD
Rv3340|M.tuberculosis_H37Rv SGQAAETFAILNLAGAGDHIVSSPRLYGGTYNLFHYSLAKLGIEVSFVDD
MMAR_1185|M.marinum_M SGQAAETFAILNLAGAGDHIVSSPRLYGGTYNLFHYSLAKLGIEVSFVDD
MUL_1438|M.ulcerans_Agy99 SGQAAETFGILNLAGAGDHIVSSPRLYGGTYNLFHYSLAKLGIEVSFVDD
MAV_4315|M.avium_104 SGQAAETFAILNLASAGDHIVSSPRLYGGTYNLFHYSLAKLGIEVSFVED
MAB_3687|M.abscessus_ATCC_1997 SGQAAATLAIVNIAQAGDNIVSSPRLYGGTYNLLHYTLPRLGISTTFVED
MSMEG_0239|M.smegmatis_MC2_155 SGLSAQFAVFAALAGAGDHIVSAASLYGGTVTQLDVSLRRFGIDTTFVGG
MLBr_02394|M.leprae_Br4923 SGMAASDCALRVMLRPGDHVIIPDDVYGGTFRLIDKVFTQWNVDYTPVP-
** :* : : **::: . :**** :. : : .:. : *
Mflv_4875|M.gilvum_PYR-GCK PDDLESWRAASRPNTKAFFAETISNPKIDVLDIPGVSGVAHDIGVPLIVD
Mvan_1554|M.vanbaalenii_PYR-1 PDDLESWRAAARPNTKAFFGETISNPQIDILDIPGVAGVAHEVGVPLIVD
TH_1970|M.thermoresistible__bu PDDLDSWRAAVRPNTKAFFGESISNPKIDILDIPNVAAVAHEAGVPLIVD
Mb3372|M.bovis_AF2122/97 PDDLDTWQAAVRPNTKAFFAETISNPQIDLLDTPAVSEVAHRNGVPLIVD
Rv3340|M.tuberculosis_H37Rv PDDLDTWQAAVRPNTKAFFAETISNPQIDLLDTPAVSEVAHRNGVPLIVD
MMAR_1185|M.marinum_M PDNLDSWQAAVRPNTKAFFGETISNPQIDLLDTPGVAEVAHRNGIPLIVD
MUL_1438|M.ulcerans_Agy99 PDNLDSWQAAVRPNTKAFFGETISNPQIDLLDTPGVAEVAHRNGIPLIVD
MAV_4315|M.avium_104 PDDLDSWQAAVRPNTKAFFAETISNPQIDILDIPGVSGVAHANGVPLIVD
MAB_3687|M.abscessus_ATCC_1997 PDDLESWRNAITPDTKALFAESISNPQNDILDIAGLSEIAHAAGIPLIVD
MSMEG_0239|M.smegmatis_MC2_155 TDPAD-YAAAVTDRTRLIYAEVISNPSGEIADLAGLAEVARDAGVPLIVD
MLBr_02394|M.leprae_Br4923 LSDLDAVRAAITSRTRLIWVETPTNPLLSIADITSIGELGKKHSVKVLVD
. : * *: :: * :** .: * . :. :.: .: ::**
Mflv_4875|M.gilvum_PYR-GCK NTIATPYLIQPLSHGADIVVHSATKYLGGHGTAIAGVIVDGGTFDWTASG
Mvan_1554|M.vanbaalenii_PYR-1 NTIATPYLIQPLSHGADIVVHSATKYLGGHGSAIAGVIVDGGTFDWTAGG
TH_1970|M.thermoresistible__bu NTIATPYLIQPLSHGADIVVHSATKYLGGHGAAIAGVIVDGGTFDWTN-G
Mb3372|M.bovis_AF2122/97 NTIATPYLIQPLAQGADIVVHSATKYLGGHGAAIAGVIVDGGNFDWTQ-G
Rv3340|M.tuberculosis_H37Rv NTIATPYLIQPLAQGADIVVHSATKYLGGHGAAIAGVIVDGGNFDWTQ-G
MMAR_1185|M.marinum_M NTIATPYLIRPFTQGADIVVHSATKYLGGHGAAIAGVIVDGGTFDWTQ-G
MUL_1438|M.ulcerans_Agy99 NTIATPYLIRPFTQGADIVVHSATKYLGGHGAAIAGVIVDGGTFDWTQ-G
MAV_4315|M.avium_104 NTIATPYLIQPFAHGADIVVHSATKYLGGHGSAIAGVIVDGGTFDWTQ-G
MAB_3687|M.abscessus_ATCC_1997 NTVATPYLIQPLAHGADVVVHSATKYLGGHGNAIGGVIIDGGTFDWTQ-G
MSMEG_0239|M.smegmatis_MC2_155 STVATPALCRPIEHGADIVVHSATKFIGGHGTTLGGVAVDAGRFDWGA-G
MLBr_02394|M.leprae_Br4923 NTFASPALQQPLMLGADVVLHSTTKYIGGHSDVVGGALVTND--------
.*.*:* * :*: ***:*:**:**::***. .:.*. : .
Mflv_4875|M.gilvum_PYR-GCK RFPGFTEPDPSYHGVVFA-ELGPPAYALKARVQLLRDLGSAAAPFNAFLI
Mvan_1554|M.vanbaalenii_PYR-1 RFPGFTSPDPSYHGVVFA-ELGPPAYALKARVQLLRDLGSAASPFNAFLI
TH_1970|M.thermoresistible__bu KFPGFTEPDPSYHGVVFA-ELGPPAFALKARVQLLRDLGSAAAPFNAFLI
Mb3372|M.bovis_AF2122/97 RFPGFTTPDPSYHGVVFA-ELGPPAFALKARVQLLRDYGSAASPFNAFLV
Rv3340|M.tuberculosis_H37Rv RFPGFTTPDPSYHGVVFA-ELGPPAFALKARVQLLRDYGSAASPFNAFLV
MMAR_1185|M.marinum_M RFPEFTTPDPSYHGVVFA-ELGAPAYALKARVQLLRDLGSAASPFNAFLV
MUL_1438|M.ulcerans_Agy99 RFPEFTTPDPSYHGVVFA-ELGAPAYALKARVQLLRDLGSAASPFNAFLV
MAV_4315|M.avium_104 RFPGFTTPDPSYHGVVFA-ELGPPAYALKARVQLLRDLGSAAAPFNAFLV
MAB_3687|M.abscessus_ATCC_1997 RHPGFTTPDPSYHGVVFA-DLGAPAYALKARVQLLRDLGPAISPFNAFLI
MSMEG_0239|M.smegmatis_MC2_155 KFPQMTEPVASYGGLSWWGNFGEFGFLTKLRSEQLRDLGATLAPQSAFQL
MLBr_02394|M.leprae_Br4923 -----EELDQAFG-------------------FLQNGAGAVPSPFDAYLT
:: .. *.. :* .*:
Mflv_4875|M.gilvum_PYR-GCK AQGLETLSLRIERHVQNAQRVAEFLEAHPDVVSVNYAGLPSSPWYGLGKK
Mvan_1554|M.vanbaalenii_PYR-1 AQGLETLSLRIERHVQNARQVAEFLDAHPDVVSVNYAGLPSSPWYDLGKK
TH_1970|M.thermoresistible__bu AQGLETLSLRVERHVENAQKVAEYLEAHPDVVSVNYAGLPSSPWYDLGKK
Mb3372|M.bovis_AF2122/97 AQGLETLSLRIERHVANAQRVAEFLAARDDVLSVNYAGLPSSPWHERAKR
Rv3340|M.tuberculosis_H37Rv AQGLETLSLRIERHVANAQRVAEFLAARDDVLSVNYAGLPSSPWHERAKR
MMAR_1185|M.marinum_M AQGLETLSLRIERHVSNAQRVAEFLADREDVVTVNYAGLPGSPWHERAKK
MUL_1438|M.ulcerans_Agy99 AQGLETLSLRIERHVSNAQRVAEFLADREDVVTVNYAGLPGSPWHERAKK
MAV_4315|M.avium_104 AQGIETLSLRMERHVANAQRVAEFLASHDGVLSVNYAGLPGSPWHERAKK
MAB_3687|M.abscessus_ATCC_1997 AQGLETLSLRVERHVANAQRVAEFLVAQPEVTRVNYAGLPTSPWYERAKA
MSMEG_0239|M.smegmatis_MC2_155 LQGVQTLPQRMAAHIANARAVAEWLEADPRVTYVRWAGLPSHPHHDRARH
MLBr_02394|M.leprae_Br4923 MRGLKTLVLRMQRHNENAITVAEFLAGHPSVSAVLYPGLPSHPGHEVAAR
:*::** *: * ** ***:* * * :.*** * : .
Mflv_4875|M.gilvum_PYR-GCK LAPKGTGAVLAFELSGGIDAGKAFVNALSLHSHVANIGDVRSLVIHPAST
Mvan_1554|M.vanbaalenii_PYR-1 LAPKGTGAVLAFELAGGVEAGKAFVNALTLHSHVANIGDVRSLVIHPAST
TH_1970|M.thermoresistible__bu LAPKGTGAVLAFELAGGVEAGKAFVNALTLHSHVANIGDVRSLVIHPAST
Mb3372|M.bovis_AF2122/97 LAPKGTGAVLSFELAGGIEAGKAFVNALKLHSHVANIGDVRSLVIHPAST
Rv3340|M.tuberculosis_H37Rv LAPKGTGAVLSFELAGGIEAGKAFVNALKLHSHVANIGDVRSLVIHPAST
MMAR_1185|M.marinum_M LSPKGTGAVLSFELAGGVEAGKAFVNALKLHSHVANIGDVRSLVIHPAST
MUL_1438|M.ulcerans_Agy99 LSPKGTGAVLSFELAGGVEAGKAFVNALKLHSHVANIGDVRSLVIHPAST
MAV_4315|M.avium_104 LAPKGTGAVLSFELAGGVEAGKAFVNALKLHSHVANIGDVRSLVIHPAST
MAB_3687|M.abscessus_ATCC_1997 LAPKGAGAVLAFELDGGVEAATTFIDSLTLHSQVANIGDVRSLVIHPATT
MSMEG_0239|M.smegmatis_MC2_155 YLPDGPGAVFAFGVKGGRAAGEKFINSVQLASHLANIGDVRTLVVHPAST
MLBr_02394|M.leprae_Br4923 QMR-GFGGMVSLRMRAGRLAAQDLCARTKVFTLAESLGGVESLIEQPSAM
* *.:.:: : .* *. : : : .:*.*.:*: :*::
Mflv_4875|M.gilvum_PYR-GCK THQQLSPEEQLASGVTPGLVRLAVGIEGIDDILADLELGFAAAKPSAAAT
Mvan_1554|M.vanbaalenii_PYR-1 THQQLSAEEQLASGVTPGLVRLAVGIEGIEDILTDLERGFAAAKPFSAAA
TH_1970|M.thermoresistible__bu THAQLSPEDQLATGVTPGLVRLAVGIEGIDDIIADLEQGFAAAKPFTASA
Mb3372|M.bovis_AF2122/97 THAQLSPAEQLATGVSPGLVRLAVGIEGIDDILADLELGFAAARRFSA--
Rv3340|M.tuberculosis_H37Rv THAQLSPAEQLATGVSPGLVRLAVGIEGIDDILADLELGFAAARRFSA--
MMAR_1185|M.marinum_M THAQLSPAEQLSTGVSPGLVRLAVGIEGIEDILADLELGFAAARKFSG--
MUL_1438|M.ulcerans_Agy99 THAQLSPAEQLSTGVSPGLVRLAVGIEGIEDILADLELGFAAAREFSG--
MAV_4315|M.avium_104 THAQLSPAEQLSTGVSPGLVRLAVGIEGIDDILADLELGFAAAAKFGANA
MAB_3687|M.abscessus_ATCC_1997 THGQLSPAEQVASGVTPGLVRLAVGIEGIDDILADLESGFTAVRRLKQ--
MSMEG_0239|M.smegmatis_MC2_155 THRQLTDEQLTAGGVGPELIRISVGIEDVEDIIWDLDQALTQAEKADD--
MLBr_02394|M.leprae_Br4923 THASTTGSQLEVP---DDLVRLSVGIEDVGDLLCDLKQALN---------
** . : : *:*::****.: *:: **. .:
Mflv_4875|M.gilvum_PYR-GCK -----DPNAVASF
Mvan_1554|M.vanbaalenii_PYR-1 -----DPNTVASF
TH_1970|M.thermoresistible__bu G---GDPQTVASF
Mb3372|M.bovis_AF2122/97 -----DPQSVAAF
Rv3340|M.tuberculosis_H37Rv -----DPQSVAAF
MMAR_1185|M.marinum_M -----DSQAVAAI
MUL_1438|M.ulcerans_Agy99 -----DSQAVAAI
MAV_4315|M.avium_104 GTSGADPHAVAAF
MAB_3687|M.abscessus_ATCC_1997 ------PAAVASS
MSMEG_0239|M.smegmatis_MC2_155 -------------
MLBr_02394|M.leprae_Br4923 -------------